Review our publications organized by year. Click here to search through our publications.
-

Evolution-guided tolerance engineering of Pseudomonas putida KT2440 for production of the aviation fuel precursor isoprenol
Lim HG, Srinivasan A, Menchavez R, Yunus IS, Noh MH, White M, Chen Y, Gin JW, Palsson BO, Lee TS, Petzold CJ, Eng T, Mukhopadhyay A, Feist AM. Evolution-guided tolerance engineering of Pseudomonas putida KT2440 for production of the aviation fuel precursor isoprenol. Metab Eng. 2025 May 19;91:322-335. doi: 10.1016/j.ymben.2025.05.007. Epub ahead of print. PMID: 40398593.
-

CRISPRi screening reveals E. coli's anaerobic-like respiratory adaptations to gentamicin: membrane depolarization by CpxR
Choe D, Lee E, Song Y, Kim SC, Jeong KJ, Palsson B, Cho B-K, Cho S. CRISPRi screening reveals E. coli's anaerobic-like respiratory adaptations to gentamicin: membrane depolarization by CpxR. mSystems. 2025 Jun 16:e0035325. doi: 10.1128/msystems.00353-25. Epub ahead of print. PMID: 40521885.
-

Regulatory orchestration of FK506 biosynthesis in Streptomyces tsukubaensisNRRL 18488 revealed through systematic analysis
Lee N, Kim W, Kim JH, Lee Y, Hwang S, Kim G, Kim H, Dan Q, Schmidt M, Yoon YJ, Cho S, Palsson B, Keasling JD, Cho BK. Regulatory orchestration of FK506 biosynthesis in Streptomyces tsukubaensisNRRL 18488 revealed through systematic analysis. iScience. 2025 May 19;28(6):112698. doi: 10.1016/j.isci.2025.112698. PMID: 40524956; PMCID: PMC12167820.
-

Trade-off between resistance and persistence in high cell density cultures
Beulig F, Bafna-Rührer J, Jensen PE, Kim SH, Patel A, Kandasamy V, Steffen C, Decker K, Zielinski D, Yang L, Özdemir E, Sudarsan S, Palsson B. Trade-off between resistance and persistence in high cell density cultures. mSystems. 2025 Jun 13:e0032325. doi: 10.1128/msystems.00323-25. Epub ahead of print. PMID: 40511936.
-

Multi-scale reactor designs extend the physical limits of CO2 fixation
Amir Akbari, Bernhard O. Palsson, Multi-scale reactor designs extend the physical limits of CO2 fixation, Chemical Engineering Journal, Volume 516, 2025, 163449,
-

Rapid identification of key antibiotic resistance genes in E. coli using high-resolution genome-scale CRISPRi screening
Rapid identification of key antibiotic resistance genes in E. coli using high-resolution genome-scale CRISPRi screening Choe, Donghui et al. iScience, Volume 28, Issue 5, 112435
-

A live bacteria enzyme assay for identification of human disease mutations and drug screening
Choe D, Palsson BO. A live bacteria enzyme assay for identification of human disease mutations and drug screening. Nat Biomed Eng. 2025 Apr 30. doi: 10.1038/s41551-025-01391-y. Epub ahead of print. PMID: 40307426.
-

Interpreting roles of mutations associated with the emergence of S. aureus USA300 strains using transcriptional regulatory network reconstruction
Poudel S, Hyun J, Hefner Y, Monk J, Nizet V, Palsson BO. Interpreting roles of mutations associated with the emergence of S. aureus USA300 strains using transcriptional regulatory network reconstruction. Elife. 2025 Apr 30;12:RP90668. doi: 10.7554/eLife.90668. PMID: 40305082; PMCID: PMC12043316.
-
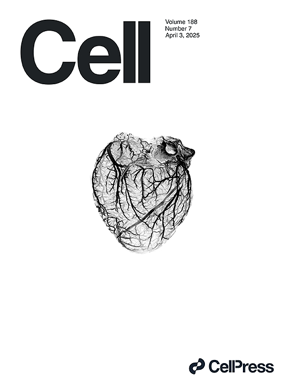
Extracellular respiration is a latent energy metabolism in Escherichia coli.
Kundu BB, Krishnan J, Szubin R, Patel A, Palsson BO, Zielinski DC, Ajo-Franklin CM. Extracellular respiration is a latent energy metabolism in Escherichia coli. Cell. 2025 Apr 4:S0092-8674(25)00289-2. doi: 10.1016/j.cell.2025.03.016. Epub ahead of print. PMID: 40215961.
-

RBC-GEM: A genome-scale metabolic model for systems biology of the human red blood cell
Haiman ZB, Key A, D'Alessandro A, Palsson BO. RBC-GEM: A genome-scale metabolic model for systems biology of the human red blood cell. PLoS Comput Biol. 2025 Mar 12;21(3):e1012109. doi: 10.1371/journal.pcbi.1012109. Epub ahead of print. PMID: 40072998.
-

Machine learning uncovers the transcriptional regulatory network for the production host Streptomyces albidoflavus
Jönsson M, Sigrist R, Gren T, Semenov Petrov M, Marcussen NEJ, Svetlova A, Charusanti P, Gockel P, Palsson BO, Yang L, Özdemir E. Machine learning uncovers the transcriptional regulatory network for the production host Streptomyces albidoflavus. Cell Rep. 2025 Mar 7;44(3):115392. doi: 10.1016/j.celrep.2025.115392. Epub ahead of print. PMID: 40057950.
-

Metabolic growth-coupling strategies for in vivo enzyme selection systems
Alter TB, Pieters PA, Lloyd CJ, Feist AM, Özdemir E, Palsson BO, Zielinski DC. Metabolic growth-coupling strategies for in vivo enzyme selection systems. Metab Eng Commun. 2025 Feb 12;20:e00257. doi: 10.1016/j.mec.2025.e00257. PMID: 40070513; PMCID: PMC11894327.
-

Pangenome mining of the Streptomyces genus redefines species' biosynthetic potential
Mohite OS, Jørgensen TS, Booth TJ, Charusanti P, Phaneuf PV, Weber T, Palsson BO. Pangenome mining of the Streptomyces genus redefines species' biosynthetic potential. Genome Biol. 2025 Jan 14;26(1):9. doi: 10.1186/s13059-024-03471-9. PMID: 39810189; PMCID: PMC11734326.
-

Decomposition of the pangenome matrix reveals a structure in gene distribution in the Escherichia coli species
Chauhan SM, Ardalani O, Hyun JC, Monk JM, Phaneuf PV, Palsson BO. Decomposition of the pangenome matrix reveals a structure in gene distribution in the Escherichia coli species. mSphere. 2024 Dec 31:e0053224. doi: 10.1128/msphere.00532-24. Epub ahead of print. PMID: 39745367.
-

Revealing systematic changes in the transcriptome during the transition from exponential growth to stationary phase.
Lim HG, Gao Y, Rychel K, Lamoureux C, Lou XA, Palsson BO. Revealing systematic changes in the transcriptome during the transition from exponential growth to stationary phase. mSystems. 2024 Dec 26:e0131524. doi: 10.1128/msystems.01315-24. Epub ahead of print. PMID: 39714213.
-

PanKB: An interactive microbial pangenome knowledgebase for research, biotechnological innovation, and knowledge mining
Sun B, Pashkova L, Pieters PA, Harke AS, Mohite OS, Santos A, Zielinski DC, Palsson BO, Phaneuf PV. PanKB: An interactive microbial pangenome knowledgebase for research, biotechnological innovation, and knowledge mining. Nucleic Acids Res. 2024 Nov 22:gkae1042. doi: 10.1093/nar/gkae1042. Epub ahead of print. PMID: 39574409.
-

Diversity of transcriptional regulatory adaptation in E. coli
Dalldorf C, Hefner Y, Szubin R, Johnsen J, Mohamed E, Li G, Krishnan J, Feist AM, Palsson BO, Zielinski DC. Diversity of transcriptional regulatory adaptation in E. coli. Mol Biol Evol. 2024 Nov 12:msae240. doi: 10.1093/molbev/msae240. Epub ahead of print. PMID: 39531644.
-

iModulonDB 2.0: dynamic tools to facilitate knowledge-mining and user-enabled analyses of curated transcriptomic datasets
Catoiu EA, Krishnan J, Li G, Lou XA, Rychel K, Yuan Y, Bajpe H, Patel A, Choe D, Shin J, Burrows J, Phaneuf PV, Zielinski DC, Palsson BO. iModulonDB 2.0: dynamic tools to facilitate knowledge-mining and user-enabled analyses of curated transcriptomic datasets. Nucleic Acids Res. 2024 Nov 4:gkae1009. doi: 10.1093/nar/gkae1009. Epub ahead of print. PMID: 39494532.
-

iModulonMiner and PyModulon: Software for unsupervised mining of gene expression compendia.
Sastry AV, Yuan Y, Poudel S, Rychel K, Yoo R, Lamoureux CR, Li G, Burrows JT, Chauhan S, Haiman ZB, Al Bulushi T, Seif Y, Palsson BO, Zielinski DC. iModulonMiner and PyModulon: Software for unsupervised mining of gene expression compendia. PLoS Comput Biol. 2024 Oct 23;20(10):e1012546. doi: 10.1371/journal.pcbi.1012546. Epub ahead of print. PMID: 39441835.
-

Diagnosis and mitigation of the systemic impact of genome reduction in Escherichia coli DGF-298
Champie A, Lachance J-C, Sastry A, Matteau D, Lloyd CJ, Grenier F, Lamoureux CR, Jeanneau S, Feist AM, Jacques P-É, Palsson BO, Rodrigue S. Diagnosis and mitigation of the systemic impact of genome reduction in Escherichia coli DGF-298. mBio. 2024 Aug 29:e0087324. doi: 10.1128/mbio.00873-24. Epub ahead of print. PMID: 39207109.
-

A genome-scale metabolic model of a globally disseminated hyperinvasive M1 strain of Streptococcus pyogenes
Hirose Y, Zielinski DC, Poudel S, Rychel K, Baker JL, Toya Y, Yamaguchi M, Heinken A, Thiele I, Kawabata S, Palsson BO, Nizet V. A genome-scale metabolic model of a globally disseminated hyperinvasive M1 strain of Streptococcus pyogenes. mSystems. 2024 Sep 17;9(9):e0073624. doi: 10.1128/msystems.00736-24. Epub 2024 Aug 19. PMID: 39158303; PMCID: PMC11406949.
-

CyuR is a dual regulator for L-cysteine dependent antimicrobial resistance in Escherichia coli
Rodionova IA, Lim HG, Gao Y, Rodionov DA, Hutchison Y, Szubin R, Dalldorf C, Monk J, Palsson BO. CyuR is a dual regulator for L-cysteine dependent antimicrobial resistance in Escherichia coli. Commun Biol. 2024 Sep 17;7(1):1160. doi: 10.1038/s42003-024-06831-0. PMID: 39289465; PMCID: PMC11408624.
-

Genome-scale models in human metabologenomics
Mardinoglu A, Palsson BØ. Genome-scale models in human metabologenomics. Nat Rev Genet. 2024 Sep 19. doi: 10.1038/s41576-024-00768-0. Epub ahead of print. PMID: 39300314.
-

Machine learning reveals the transcriptional regulatory network and circadian dynamics of Synechococcus elongatusPCC 7942
Yuan Y, Al Bulushi T, Sastry AV, Sancar C, Szubin R, Golden SS, Palsson BO. Machine learning reveals the transcriptional regulatory network and circadian dynamics of Synechococcus elongatusPCC 7942. Proc Natl Acad Sci U S A. 2024 Sep 17;121(38):e2410492121. doi: 10.1073/pnas.2410492121. Epub 2024 Sep 13. PMID: 39269777.
-

Machine-Learning Analysis of Streptomyces coelicolor Transcriptomes Reveals a Transcription Regulatory Network Encompassing Biosynthetic Gene Clusters
Lee Y, Choe D, Palsson BO, Cho BK. Machine-Learning Analysis of Streptomyces coelicolor Transcriptomes Reveals a Transcription Regulatory Network Encompassing Biosynthetic Gene Clusters. Adv Sci (Weinh). 2024 Sep 12:e2403912. doi: 10.1002/advs.202403912. Epub ahead of print. PMID: 39264300.
-

Modulating bacterial function utilizing A knowledge base of transcriptional regulatory modules
Shin J, Zielinski DC, Palsson BO. Modulating bacterial function utilizing A knowledge base of transcriptional regulatory modules. Nucleic Acids Res. 2024 Aug 28:gkae742. doi: 10.1093/nar/gkae742. Epub ahead of print. PMID: 39193902.
-
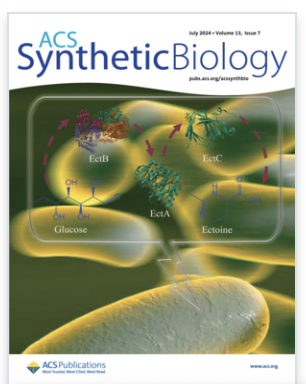
Meta-analysis Driven Strain Design for Mitigating Oxidative Stresses Important in Biomanufacturing
Phaneuf PV, Kim SH, Rychel K, Rode C, Beulig F, Palsson BO, Yang L. Meta-analysis Driven Strain Design for Mitigating Oxidative Stresses Important in Biomanufacturing. ACS Synth Biol. 2024 Jul 19;13(7):2045-2059. doi: 10.1021/acssynbio.3c00572. Epub 2024 Jun 27. PMID: 38934464; PMCID: PMC11264330.
-

Phosphofructokinase isoforms boost energy status of stored red blood cells and transfusion outcomes.
Nemkov T, Stephenson D, Earley EJ, Keele GR, Hay A, Key A, Haiman ZB, Erickson C, Dzieciatkowska M, Reisz JA, Moore A, Stone M, Deng X, Kleinman S, Spitalnik SL, Hod EA, Hudson KE, Hansen KC, Palsson BO, Churchill GA, Roubinian N, Norris PJ, Busch MP, Zimring JC, Page GP, D'Alessandro A. Biological and genetic determinants of glycolysis: Phosphofructokinase isoforms boost energy status of stored red blood cells and transfusion outcomes. Cell Metab. 2024 Jun 27:S1550-4131(24)00232-8. doi: 10.1016/j.cmet.2024.06.007. Epub ahead of print. PMID: 38964323.
-
Pangenome reconstruction of Lactobacillaceae metabolism predicts species-specific metabolic traits
Ardalani O, Phaneuf PV, Mohite OS, Nielsen LK, Palsson BO. Pangenome reconstruction of Lactobacillaceae metabolism predicts species-specific metabolic traits. mSystems. 2024 Jun 26:e0015624. doi: 10.1128/msystems.00156-24. Epub ahead of print. PMID: 38920366.
-

A treasure trove of 1034 actinomycete genomes
Jørgensen TS, Mohite OS, Sterndorff EB, Alvarez-Arevalo M, Blin K, Booth TJ, Charusanti P, Faurdal D, Hansen TØ, Nuhamunada M, Mourched AS, Palsson BØ, Weber T. A treasure trove of 1034 actinomycete genomes. Nucleic Acids Res. 2024 Jun 22:gkae523. doi: 10.1093/nar/gkae523. Epub ahead of print. PMID: 38908028.
-

Proteome allocation is linked to transcriptional regulation through a modularized transcriptome
Patel A, McGrosso D, Hefner Y, Campeau A, Sastry AV, Maurya S, Rychel K, Gonzalez DJ, Palsson BO. Proteome allocation is linked to transcriptional regulation through a modularized transcriptome. Nat Commun. 2024 Jun 19;15(1):5234. doi: 10.1038/s41467-024-49231-y. PMID: 38898010.
-
The hallmarks of a tradeoff in transcriptomes that balances stress and growth functions
Dalldorf C, Rychel K, Szubin R, Hefner Y, Patel A, Zielinski DC, Palsson BO. The hallmarks of a tradeoff in transcriptomes that balances stress and growth functions. mSystems. 2024 Jun 3:e0030524. doi: 10.1128/msystems.00305-24. Epub ahead of print. PMID: 38829048.
-

Deciphering Nutritional Stress Responses via Knowledge-Enriched Transcriptomics for Microbial Engineering
Shin J, Zielinski DC, Palsson BO. Deciphering Nutritional Stress Responses via Knowledge-Enriched Transcriptomics for Microbial Engineering. Metab Eng. 2024 May 31:S1096-7176(24)00073-9. doi: 10.1016/j.ymben.2024.05.007. Epub ahead of print. PMID: 38825177.
-

Escherichia coli non-coding regulatory regions are highly conserved
Lamoureux CR, Phaneuf PV, Palsson BO, Zielinski DC. Escherichia coli non-coding regulatory regions are highly conserved. NAR Genom Bioinform. 2024 May 20;6(2):lqae041. doi: 10.1093/nargab/lqae041. PMID: 38774514; PMCID: PMC11106028.
-

BGCFlow: systematic pangenome workflow for the analysis of biosynthetic gene clusters across large genomic datasets
Nuhamunada M, Mohite OS, Phaneuf PV, Palsson BO, Weber T. BGCFlow: systematic pangenome workflow for the analysis of biosynthetic gene clusters across large genomic datasets. Nucleic Acids Res. 2024 Apr 30:gkae314. doi: 10.1093/nar/gkae314. Epub ahead of print. PMID: 38686794.
-

Bottom-up parameterization of enzyme rate constants: Reconciling inconsistent data
Daniel C. Zielinski, Marta R.A. Matos, James E. de Bree, Kevin Glass, Nikolaus Sonnenschein, Bernhard O. Palsson, Bottom-up parameterization of enzyme rate constants: Reconciling inconsistent data, Metabolic Engineering Communications, Volume 18, 2024, e00234, ISSN 2214-0301, https://doi.org/10.1016/j.mec.2024.e00234.
-

Grand Challenges at the Interface of Engineering and Medicine
Subramaniam S, Akay M, Anastasio MA, Bailey V, Boas D, Bonato P, Chilkoti A, Cochran JR, Colvin V, Desai TA, Duncan JS, Epstein FH, Fraley S, Giachelli C, Grande-Allen KJ, Green J, Guo XE, Hilton IB, Humphrey JD, Johnson CR, Karniadakis G, King MR, Kirsch RF, Kumar S, Laurencin CT, Li S, Lieber RL, Lovell N, Mali P, Margulies SS, Meaney DF, Ogle B, Palsson B, A Peppas N, Perreault EJ, Rabbitt R, Setton LA, Shea LD, Shroff SG, Shung K, Tolias AS, van der Meulen MCH, Varghese S, Vunjak-Novakovic G, White JA, Winslow R, Zhang J, Zhang K, Zukoski C, Miller MI. Grand Challenges at the Interface of Engineering and Medicine. IEEE Open J Eng Med Biol. 2024 Feb 21;5:1-13. doi: 10.1109/OJEMB.2024.3351717. PMID: 38415197; PMCID: PMC10896418.
-

Serial adaptive laboratory evolution enhances mixed carbon metabolic capacity of Escherichia coli
Kim K, Choe D, Kang M, Cho SH, Cho S, Jeong KJ, Palsson B, Cho BK. Serial adaptive laboratory evolution enhances mixed carbon metabolic capacity of Escherichia coli. Metab Eng. 2024 Apr 16:S1096-7176(24)00058-2. doi: 10.1016/j.ymben.2024.04.004. Epub ahead of print. PMID: 38636729.
-

Reduction-to-synthesis: the dominant approach to genome-scale synthetic biology
Kim K, Choe D, Cho S, Palsson B, Cho BK. Reduction-to-synthesis: the dominant approach to genome-scale synthetic biology. Trends Biotechnol. 2024 Feb 28:S0167-7799(24)00037-4. doi: 10.1016/j.tibtech.2024.02.008. Epub ahead of print. PMID: 38423803.
-

Advancing the scale of synthetic biology via cross-species transfer of cellular functions enabled by iModulon engraftment
Choe D, Olson CA, Szubin R, Yang H, Sung J, Feist AM, Palsson BO. Advancing the scale of synthetic biology via cross-species transfer of cellular functions enabled by iModulon engraftment. Nat Commun. 2024 Mar 15;15(1):2356. doi: 10.1038/s41467-024-46486-3. PMID: 38490991; PMCID: PMC10943186.
-

Unified computing framework of Escherichia coli metabolism, gene expression, and stress responses
Zhao J, Chen K, Palsson BO, Yang L. StressME: Unified computing framework of Escherichia coli metabolism, gene expression, and stress responses. PLoS Comput Biol. 2024 Feb 12;20(2):e1011865. doi: 10.1371/journal.pcbi.1011865. Epub ahead of print. PMID: 38346086.
-

Reconstructing the transcriptional regulatory network of probiotic L. reuteri is enabled by transcriptomics and machine learning
Josephs-Spaulding J, Rajput A, Hefner Y, Szubin R, Balasubramanian A, Li G, Zielinski DC, Jahn L, Sommer M, Phaneuf P, Palsson BO. Reconstructing the transcriptional regulatory network of probiotic L. reuteri is enabled by transcriptomics and machine learning. mSystems. 2024 Feb 13:e0125723. doi: 10.1128/msystems.01257-23. Epub ahead of print. PMID: 38349131.
-

Machine learning analysis of RB-TnSeq fitness data predicts functional gene modules in Pseudomonas putida KT2440
Borchert AJ, Bleem AC, Lim HG, Rychel K, Dooley KD, Kellermyer ZA, Hodges TL, Palsson BO, Beckham GT. Machine learning analysis of RB-TnSeq fitness data predicts functional gene modules in Pseudomonas putida KT2440. mSystems. 2024 Feb 7:e0094223. doi: 10.1128/msystems.00942-23. Epub ahead of print. PMID: 38323821.
-

Inferred regulons are consistent with regulator binding sequences in E. coli.
Qiu S, Wan X, Liang Y, Lamoureux CR, Akbari A, Palsson BO, Zielinski DC. Inferred regulons are consistent with regulator binding sequences in E. coli. PLoS Comput Biol. 2024 Jan 22;20(1):e1011824. doi: 10.1371/journal.pcbi.1011824. PMID: 38252668; PMCID: PMC10833566.
-

A data-driven approach for timescale decomposition of biochemical reaction networks
Akbari A, Haiman ZB, Palsson BO. A data-driven approach for timescale decomposition of biochemical reaction networks. mSystems. 2024 Jan 23:e0100123. doi: 10.1128/msystems.01001-23. Epub ahead of print. PMID: 38259168.
-

Independent component analysis reveals 49 independently modulated gene sets within the global transcriptional regulatory architecture of multidrug-resistant Acinetobacter baumannii.
Menon ND, Poudel S, Sastry AV, Rychel K, Szubin R, Dillon N, Tsunemoto H, Hirose Y, Nair BG, Kumar GB, Palsson BO, Nizet V. Independent component analysis reveals 49 independently modulated gene sets within the global transcriptional regulatory architecture of multidrug-resistant Acinetobacter baumannii. mSystems. 2024 Jan 8:e0060623. doi: 10.1128/msystems.00606-23. Epub ahead of print. PMID: 38189271.
-

Experimental promoter identification of a foodborne pathogen Salmonella enterica subsp. enterica serovar Typhimurium with near single base-pair resolution
Lee Sang-Mok, Le Hoa Thi, Taizhanova Assiya, Nong Linh Khanh, Park Joon Young, Lee Eun-Jin, Palsson Bernhard O., Kim Donghyuk, "Experimental promoter identification of a foodborne pathogen Salmonella enterica subsp. enterica serovar Typhimurium with near single base-pair resolution," Frontiers in Microbiology 14. 2024
-

Global pathogenomic analysis identifies known and candidate genetic antimicrobial resistance determinants in twelve species
Hyun JC, Monk JM, Szubin R, Hefner Y, Palsson BO. Global pathogenomic analysis identifies known and candidate genetic antimicrobial resistance determinants in twelve species. Nat Commun. 2023 Nov 24;14(1):7690. doi: 10.1038/s41467-023-43549-9. PMID: 38001096; PMCID: PMC10673929.
-

High-resolution temporal profiling of E. coli transcriptional response
Miano A, Rychel K, Lezia A, Sastry A, Palsson B, Hasty J. High-resolution temporal profiling of E. coli transcriptional response. Nat Commun. 2023 Nov 22;14(1):7606. doi: 10.1038/s41467-023-43173-7. PMID: 37993418; PMCID: PMC10665441.
-

Functional annotation of enzyme-encoding genes using deep learning with transformer layers
Kim GB, Kim JY, Lee JA, Norsigian CJ, Palsson BO, Lee SY. Functional annotation of enzyme-encoding genes using deep learning with transformer layers. Nat Commun. 2023 Nov 14;14(1):7370. doi: 10.1038/s41467-023-43216-z. PMID: 37963869; PMCID: PMC10645960.
-

Modeling Red Blood Cell Metabolism in the Omics Era
Key A, Haiman Z, Palsson BO, D’Alessandro A. Modeling Red Blood Cell Metabolism in the Omics Era. Metabolites. 2023; 13(11):1145. https://doi.org/10.3390/metabo13111145
-

Differential Expression Analysis Utilizing Condition-Specific Metabolic Pathways
Mattei, G.; Gan, Z.; Ramazzotti, M.; Palsson, B.O.; Zielinski, D.C. Differential Expression Analysis Utilizing Condition-Specific Metabolic Pathways. Metabolites 2023, 13, 1127. https://doi.org/10.3390/metabo13111127
-

Laboratory evolution, transcriptomics, and modeling reveal mechanisms of paraquat tolerance
Rychel K, Tan J, Patel A, Lamoureux C, Hefner Y, Szubin R, Johnsen J, Mohamed ETT, Phaneuf PV, Anand A, Olson CA, Park JH, Sastry AV, Yang L, Feist AM, Palsson BO. Laboratory evolution, transcriptomics, and modeling reveal mechanisms of paraquat tolerance. Cell Rep. 2023 Sep 13;42(9):113105. doi: 10.1016/j.celrep.2023.113105. Epub ahead of print. PMID: 37713311.
-

A multi-scale expression and regulation knowledge base for Escherichia coli
Lamoureux CR, Decker KT, Sastry AV, Rychel K, Gao Y, McConn JL, Zielinski DC, Palsson BO. A multi-scale expression and regulation knowledge base for Escherichia coli. Nucleic Acids Res. 2023 Sep 15:gkad750. doi: 10.1093/nar/gkad750. Epub ahead of print. PMID: 37713610.
-

Recent advances in non-model bacterial chassis construction
Hwang S, Joung C, Kim W, Palsson B, Cho B-K, Recent advances in non-model bacterial chassis construction, Current Opinion in Systems Biology, https://doi.org/10.1016/ j.coisb.2023.100471.
-

Machine learning uncovers the Pseudomonas syringae transcriptome in microbial communities and during infection
Bajpe H, Rychel K, Lamoureux CR, Sastry AV, Palsson BO. Machine learning uncovers the Pseudomonas syringae transcriptome in microbial communities and during infection. mSystems. 2023 Aug 28:e0043723. doi: 10.1128/msystems.00437-23. Epub ahead of print. PMID: 37638727.
-

Reconstruction of the last bacterial common ancestor from 183 pangenomes reveals a versatile ancient core genome
Hyun, J.C., Palsson, B.O. Reconstruction of the last bacterial common ancestor from 183 pangenomes reveals a versatile ancient core genome. Genome Biol 24, 183 (2023). https://doi.org/10.1186/s13059-023-03028-2
-

Adaptive Evolution of a Minimal Organism with a Synthetic Genome
Sandberg, T.E., Wise, K.S., Dalldorf, C., Szubin, R., Feist, A.M., Glass, J.I., Palsson, B.O., Adaptive Evolution of a Minimal Organism with a Synthetic Genome ISCIENCE (2023), doi: https://doi.org/10.1016/j.isci.2023.107500.
-

Pangenome analysis reveals the genetic basis for taxonomic classification of the Lactobacillaceae family
Akanksha Rajput, Siddharth M. Chauhan, Omkar S. Mohite, Jason C. Hyun, Omid Ardalani, Leonie J. Jahn, Morten OA. Sommer, Bernhard O. Palsson, Pangenome analysis reveals the genetic basis for taxonomic classification of the Lactobacillaceae family, Food Microbiology, Volume 115, 2023
-

Elucidating the CodY regulon in Staphylococcus aureus USA300 substrains TCH1516 and LAC
Gao Y, Poudel S, Seif Y, Shen Z, Palsson BO. Elucidating the CodY regulon in Staphylococcus aureus USA300 substrains TCH1516 and LAC. mSystems. 2023 Jun 13:e0027923. doi: 10.1128/msystems.00279-23. Epub ahead of print. PMID: 37310465.
-

Empowering drug off-target discovery with metabolic and structural analysis
Chowdhury S, Zielinski DC, Dalldorf C, Rodrigues JV, Palsson BO, Shakhnovich EI. Empowering drug off-target discovery with metabolic and structural analysis. Nat Commun. 2023 Jun 9;14(1):3390. doi: 10.1038/s41467-023-38859-x. PMID: 37296102; PMCID: PMC10256842.
-

Elucidation of independently modulated genes in Streptococcus pyogenes reveals carbon sources that control its expression of hemolytic toxins
Hirose Y, Poudel S, Sastry AV, Rychel K, Lamoureux CR, Szubin R, Zielinski DC, Lim HG, Menon ND, Bergsten H, Uchiyama S, Hanada T, Kawabata S, Palsson BO, Nizet V. Elucidation of independently modulated genes in Streptococcus pyogenes reveals carbon sources that control its expression of hemolytic toxins. mSystems. 2023 Jun 6:e0024723. doi: 10.1128/msystems.00247-23. Epub ahead of print. PMID: 37278526.
-

E. coli allantoinase is activated by the downstream metabolic enzyme, glycerate kinase, and stabilizes the putative allantoin transporter by direct binding
Rodionova IA, Hosseinnia A, Kim S, Goodacre N, Zhang L, Zhang Z, Palsson B, Uetz P, Babu M, Saier MH Jr. E. coli allantoinase is activated by the downstream metabolic enzyme, glycerate kinase, and stabilizes the putative allantoin transporter by direct binding. Sci Rep. 2023 May 5;13(1):7345. doi: 10.1038/s41598-023-31812-4. PMID: 37147430; PMCID: PMC10163214.
-

Systems biology of competency in Vibrio natriegens is revealed by applying novel data analytics to the transcriptome
Shin J, Rychel K, Palsson BO. Systems biology of competency in Vibrio natriegens is revealed by applying novel data analytics to the transcriptome. Cell Rep. 2023 Jun 6;42(6):112619. doi: 10.1016/j.celrep.2023.112619. Epub ahead of print. PMID: 37285268.
-

Metabolic homeostasis and growth in abiotic cells
Akbari A, Palsson BO. Metabolic homeostasis and growth in abiotic cells. Proc Natl Acad Sci U S A. 2023 May 9;120(19):e2300687120. doi: 10.1073/pnas.2300687120. Epub 2023 May 1. PMID: 37126695.
-

The Escherichia coli Fur pan-regulon has few conserved but many unique regulatory targets
Gao Y, Bang I, Seif Y, Kim D, Palsson BO. The Escherichia coli Fur pan-regulon has few conserved but many unique regulatory targets. Nucleic Acids Res. 2023 Apr 7:gkad253. doi: 10.1093/nar/gkad253. Epub ahead of print. PMID: 37026477.
-

Whole-genome sequences from wild-type and laboratory-evolved strains define the alleleome and establish its hallmarks
Catoiu EA, Phaneuf P, Monk J, Palsson BO. Whole-genome sequences from wild-type and laboratory-evolved strains define the alleleome and establish its hallmarks. Proc Natl Acad Sci U S A. 2023;120(15):e2218835120. doi:10.1073/pnas.2218835120
-

A model industrial workhorse: Bacillus subtilis strain 168 and its genome after a quarter of a century
Bremer E, Calteau A, Danchin A, Harwood C, Helmann JD, Médigue C, Palsson BO, Sekowska A, Vallenet D, Zuniga A, Zuniga C. A model industrial workhorse: Bacillus subtilis strain 168 and its genome after a quarter of a century. Microb Biotechnol. 2023 Apr 1. doi: 10.1111/1751-7915.14257. Epub ahead of print. PMID: 37002859.
-

Revealing oxidative pentose metabolism in new Pseudomonas putida isolates
Park, M. R., Gauttam, R., Fong, B., Chen, Y., Lim, H. G., Feist, A. M., Mukhopadhyay, A., Petzold, C. J., Simmons, B. A., & Singer, S. W. (2023). Revealing oxidative pentose metabolism in new Pseudomonas putida isolates. Environmental microbiology, 25(2), 493–504. https://doi.org/10.1111/1462-2920.16296
-

Laboratory evolution reveals general and specific tolerance mechanisms for commodity chemicals
Lennen RM, Lim HG, Jensen K, Mohammed ET, Phaneuf PV, Noh MH, Malla S, Börner RA, Chekina K, Özdemir E, Bonde I, Koza A, Maury J, Pedersen LE, Schöning LY, Sonnenschein N, Palsson BO, Nielsen AT, Sommer MOA, Herrgård MJ, Feist AM. Laboratory evolution reveals general and specific tolerance mechanisms for commodity chemicals. Metab Eng. 2023 Feb 2:S1096-7176(23)00020-4. doi: 10.1016/j.ymben.2023.01.012. Epub ahead of print. PMID: 36738854.
-

Deep-learning optimized DEOCSU suite provides an iterable pipeline for accurate ChIP-exo peak calling
Bang I, Lee SM, Park S, Park JY, Nong LK, Gao Y, Palsson BO, Kim D. Deep-learning optimized DEOCSU suite provides an iterable pipeline for accurate ChIP-exo peak calling. Brief Bioinform. 2023 Jan 25:bbad024. doi: 10.1093/bib/bbad024. Epub ahead of print. PMID: 36702751.
-

Model-driven experimental design workflow expands understanding of regulatory role of Nac in Escherichia coli
Park JY, Lee SM, Ebrahim A, Scott-Nevros ZK, Kim J, Yang L, Sastry A, Seo SW, Palsson BO, Kim D. Model-driven experimental design workflow expands understanding of regulatory role of Nac in Escherichia coli. NAR Genom Bioinform. 2023 Jan 20;5(1):lqad006. doi: 10.1093/nargab/lqad006. PMID: 36685725; PMCID: PMC9853098.
-

Comprehensive whole genome sequencing with hybrid assembly of multi-drug resistant Candida albicans isolate causing cerebral abscess
Monika Kumaraswamy, Alison Coady, Richard Szubin, Thomas CS Martin, Bernhard Palsson, Victor Nizet, Jonathan M. Monk, Comprehensive whole genome sequencing with hybrid assembly of multi-drug resistant Candida albicans isolate causing cerebral abscess, Current Research in Microbial Sciences, Volume 4, 2023, 100180, ISSN 2666-5174, https://doi.org/10.1016/j.crmicr.2023.100180. (https://www.sciencedirect.com/science/article/pii/S2666517423000019)
-

A systems approach discovers the role and characteristics of seven LysR type transcription factors in Escherichia coli
Rodionova IA, Gao Y, Monk J, Hefner Y, Wong N, Szubin R, Lim HG, Rodionov DA, Zhang Z, Saier MH Jr, Palsson BO. A systems approach discovers the role and characteristics of seven LysR type transcription factors in Escherichia coli. Sci Rep. 2022 May 4;12(1):7274. doi: 10.1038/s41598-022-11134-7. PMID: 35508583; PMCID: PMC9068703.
-

Experimental Evolution Reveals Unifying Systems-Level Adaptations but Diversity in Driving Genotypes
Kavvas ES, Long CP, Sastry A, Poudel S, Antoniewicz MR, Ding Y, Mohamed ET, Szubin R, Monk JM, Feist AM, Palsson BO. Experimental Evolution Reveals Unifying Systems-Level Adaptations but Diversity in Driving Genotypes. mSystems. 2022 Oct 13:e0016522. doi: 10.1128/msystems.00165-22. Epub ahead of print. PMID: 36226969.
-

An unexpected role for leucyl aminopeptidase in UV tolerance revealed by a genome-wide fitness assessment in a model cyanobacterium
Weiss EL, Fang M, Taton A, Szubin R, Palsson BØ, Mitchell BG, Golden SS. An unexpected role for leucyl aminopeptidase in UV tolerance revealed by a genome-wide fitness assessment in a model cyanobacterium. Proc Natl Acad Sci U S A. 2022 Nov 8;119(45):e2211789119. doi: 10.1073/pnas.2211789119. Epub 2022 Nov 2. PMID: 36322730.
-

Coordination of CcpA and CodY Regulators in Staphylococcus aureus USA300 Strains
Poudel S, Hefner Y, Szubin R, Sastry A, Gao Y, Nizet V, Palsson BO. Coordination of CcpA and CodY Regulators in Staphylococcus aureus USA300 Strains. mSystems. 2022 Nov 2:e0048022. doi: 10.1128/msystems.00480-22. Epub ahead of print. PMID: 36321827.
-

Pan-Genome Analysis of Transcriptional Regulation in Six Salmonella enterica Serovar Typhimurium Strains Reveals Their Different Regulatory Structures
Yuan Y, Seif Y, Rychel K, Yoo R, Chauhan S, Poudel S, Al-Bulushi T, Palsson BO, Sastry AV. Pan-Genome Analysis of Transcriptional Regulation in Six Salmonella enterica Serovar Typhimurium Strains Reveals Their Different Regulatory Structures. mSystems. 2022 Nov 1:e0046722. doi: 10.1128/msystems.00467-22. Epub ahead of print. PMID: 36317888.
-

Revealing Causes for False-Positive and False-Negative Calling of Gene Essentiality in Escherichia coli Using Transposon Insertion Sequencing
Choe D, Kim U, Hwang S, Seo SW, Kim D, Cho S, Palsson B, Cho BK. Revealing Causes for False-Positive and False-Negative Calling of Gene Essentiality in Escherichia coli Using Transposon Insertion Sequencing. mSystems. 2022 Dec 12:e0089622. doi: 10.1128/msystems.00896-22. Epub ahead of print. PMID: 36507678.
-

High-quality genome-scale metabolic network reconstruction of probiotic bacterium Escherichia coli Nissle 1917
van 't Hof M, Mohite OS, Monk JM, Weber T, Palsson BO, Sommer MOA. High-quality genome-scale metabolic network reconstruction of probiotic bacterium Escherichia coli Nissle 1917. BMC Bioinformatics. 2022 Dec 30;23(1):566. doi: 10.1186/s12859-022-05108-9. PMID: 36585633; PMCID: PMC9801561.
-

Minireview: Engineering evolution to reconfigure phenotypic traits in microbes for biotechnological applications Add to Default shortcuts
Kangsan Kim, Minjeong Kang, Sang-Hyeok Cho, Eojin Yoo, Ui-Gi Kim, Suhyung Cho, Bernhard Palsson, Byung-Kwan Cho, Minireview: Engineering evolution to reconfigure phenotypic traits in microbes for biotechnological applications, Computational and Structural Biotechnology Journal, Volume 21, 2023, Pages 563-573, ISSN 2001-0370, https://doi.org/10.1016/j.csbj.2022.12.042.
-

Integration of physiologically relevant photosynthetic energy flows into whole genome models of light-driven metabolism
Broddrick JT, Ware MA, Jallet D, Palsson BO, Peers G. Integration of physiologically relevant photosynthetic energy flows into whole genome models of light-driven metabolism. Plant J. 2022 Sep 2. doi: 10.1111/tpj.15965. Epub ahead of print. PMID: 36053127.
-

Advanced transcriptomic analysis reveals the role of efflux pumps and media composition in antibiotic responses of Pseudomonas aeruginosa
Rajput A, Tsunemoto H, Sastry AV, Szubin R, Rychel K, Chauhan SM, Pogliano J, Palsson BO. Advanced transcriptomic analysis reveals the role of efflux pumps and media composition in antibiotic responses of Pseudomonas aeruginosa. Nucleic Acids Res. 2022 Sep 12:gkac743. doi: 10.1093/nar/gkac743. Epub ahead of print. PMID: 36095122.
-

Positively charged mineral surfaces promoted the accumulation of organic intermediates at the origin of metabolism
Akbari A, Palsson BO. Positively charged mineral surfaces promoted the accumulation of organic intermediates at the origin of metabolism. PLoS Comput Biol. 2022 Aug 17;18(8):e1010377. doi: 10.1371/journal.pcbi.1010377. PMID: 35976973; PMCID: PMC9423644.
-

Adaptive laboratory evolution and independent component analysis disentangle complex vancomycin adaptation trajectories
Fait, A., Seif, Y., Mikkelsen, K., Poudel, S., Wells, J. M., Palsson, B. O., & Ingmer, H. (2022). Adaptive laboratory evolution and independent component analysis disentangle complex vancomycin adaptation trajectories. Proceedings of the National Academy of Sciences of the United States of America, 119(30), e2118262119. https://doi.org/10.1073/pnas.2118262119
-

Laboratory evolution of synthetic electron transport system variants reveals a larger metabolic respiratory system and its plasticity
Anand A, Patel A, Chen K, Olson CA, Phaneuf PV, Lamoureux C, Hefner Y, Szubin R, Feist AM, Palsson BO. Laboratory evolution of synthetic electron transport system variants reveals a larger metabolic respiratory system and its plasticity. Nat Commun. 2022 Jun 27;13(1):3682. doi: 10.1038/s41467-022-30877-5. PMID: 35760776.
-

Identification and Engineering of Transporters for Efficient Melatonin Production in Escherichia coli
Yang L, Malla S, Özdemir E, Kim SH, Lennen R, Christensen HB, Christensen U, Munro LJ, Herrgård MJ, Kell DB, Palsson BØ. Identification and Engineering of Transporters for Efficient Melatonin Production in Escherichia coli. Front Microbiol [Internet]. 2022;13. Available from: https://www.frontiersin.org/article/10.3389/fmicb.2022.880847
-

Pangenome analysis of Enterobacteria reveals richness of secondary metabolite gene clusters and their associated gene sets
Mohite OS, Lloyd CJ, Monk JM, Weber T, Palsson BO. Pangenome analysis of Enterobacteria reveals richness of secondary metabolite gene clusters and their associated gene sets. Synth Syst Biotechnol. 2022 May 6;7(3):900-910. doi: 10.1016/j.synbio.2022.04.011. PMID: 35647330; PMCID: PMC9125672.
-

Machine-learning from Pseudomonas putida KT2440 transcriptomes reveals its transcriptional regulatory network
Lim HG, Rychel K, Sastry AV, Bentley GJ, Mueller J, Schindel HS, Larsen PE, Laible PD, Guss AM, Niu W, Johnson CW, Beckham GT, Feist AM, Palsson BO. Machine-learning from Pseudomonas putida KT2440 transcriptomes reveals its transcriptional regulatory network. Metab Eng. 2022 Apr 27:S1096-7176(22)00063-5. doi: 10.1016/j.ymben.2022.04.004. Epub ahead of print. PMID: 35489688.
-

Systems biology approach to functionally assess the Clostridioides difficile pangenome reveals genetic diversity with discriminatory power
Norsigian CJ, Danhof HA, Brand CK, Midani FS, Broddrick JT, Savidge TC, Britton RA, Palsson BO, Spinler JK, Monk JM. Systems biology approach to functionally assess the Clostridioides difficile pangenome reveals genetic diversity with discriminatory power. Proc Natl Acad Sci U S A. 2022 May 3;119(18):e2119396119. doi: 10.1073/pnas.2119396119. Epub 2022 Apr 27. PMID: 35476524.
-

Regulatory perturbations of ribosome allocation in bacteria reshape the growth proteome with a trade-off in adaptation capacity
Hidalgo D, Martínez-Ortiz CA, Palsson BO, Jiménez JI, Utrilla J. Regulatory perturbations of ribosome allocation in bacteria reshape the growth proteome with a trade-off in adaptation capacity. iScience. 2022 Feb 7;25(3):103879. doi: 10.1016/j.isci.2022.103879. PMID: 35243241; PMCID: PMC8866900.
-

System-Level Analysis of Transcriptional and Translational Regulatory Elements in Streptomyces griseus
Hwang S, Lee N, Choe D, Lee Y, Kim W, Kim JH, Kim G, Kim H, Ahn NH, Lee BH, Palsson BO, Cho BK. System-Level Analysis of Transcriptional and Translational Regulatory Elements in Streptomyces griseus. Front Bioeng Biotechnol. 2022 Feb 25;10:844200. doi: 10.3389/fbioe.2022.844200. PMID: 35284422; PMCID: PMC8914203.
-

Synthetic 3'-UTR valves for optimal metabolic flux control in Escherichia coli
Choe D, Kim K, Kang M, Lee SG, Cho S, Palsson B, Cho BK. Synthetic 3'-UTR valves for optimal metabolic flux control in Escherichia coli. Nucleic Acids Res. 2022 Mar 31:gkac206. doi: 10.1093/nar/gkac206. Epub ahead of print. PMID: 35357499.
-

Machine learning from Pseudomonas aeruginosa transcriptomes identifies independently modulated sets of genes associated with known transcriptional regulators
Rajput A, Tsunemoto H, Sastry AV, Szubin R, Rychel K, Sugie J, Pogliano J, Palsson BO. Machine learning from Pseudomonas aeruginosa transcriptomes identifies independently modulated sets of genes associated with known transcriptional regulators. Nucleic Acids Res. 2022 Mar 31:gkac187. doi: 10.1093/nar/gkac187. Epub ahead of print. PMID: 35357493.
-

Machine Learning of All Mycobacterium tuberculosis H37Rv RNA-seq Data Reveals a Structured Interplay between Metabolism, Stress Response, and Infection
Yoo R, Rychel K, Poudel S, Al-Bulushi T, Yuan Y, Chauhan S, Lamoureux C, Palsson BO, Sastry A. Machine Learning of All Mycobacterium tuberculosis H37Rv RNA-seq Data Reveals a Structured Interplay between Metabolism, Stress Response, and Infection. mSphere. 2022 Mar 21:e0003322. doi: 10.1128/msphere.00033-22. Epub ahead of print. PMID: 35306876.
-

Is the kinetome conserved?
Palsson BO, Yurkovich JT. Is the kinetome conserved? Mol Syst Biol. 2022 Feb;18(2):e10782. doi: 10.15252/msb.202110782. PMID: 35188334.
-

Genome-scale analysis of genetic regulatory elements in Streptomyces avermitilis MA-4680 using transcript boundary information
Lee Y, Lee N, Hwang S, Kim W, Cho S, Palsson BO, Cho BK. Genome-scale analysis of genetic regulatory elements in Streptomyces avermitilis MA-4680 using transcript boundary information. BMC Genomics. 2022 Jan 21;23(1):68. doi: 10.1186/s12864-022-08314-0. PMID: 35062881.
-

Comparative pangenomics: analysis of 12 microbial pathogen pangenomes reveals conserved global structures of genetic and functional diversity
Hyun JC, Monk JM, Palsson BO. Comparative pangenomics: analysis of 12 microbial pathogen pangenomes reveals conserved global structures of genetic and functional diversity. BMC Genomics. 2022 Jan 4;23(1):7. doi: 10.1186/s12864-021-08223-8. PMID: 34983386; PMCID: PMC8725406.
-

Streptomyces as Microbial Chassis for Heterologous Protein Expression
Hwang S, Lee Y, Kim JH, Kim G, Kim H, Kim W, Cho S, Palsson BO, Cho BK. Streptomyces as Microbial Chassis for Heterologous Protein Expression. Front Bioeng Biotechnol. 2021 Dec 21;9:804295. doi: 10.3389/fbioe.2021.804295. PMID: 34993191; PMCID: PMC8724576.
-

Re-classification of Streptomyces venezuelae strains and mining secondary metabolite biosynthetic gene clusters
Lee N, Choi M, Kim W, Hwang S, Lee Y, Kim JH, Kim G, Kim H, Cho S, Kim SC, Palsson B, Jang KS, Cho BK. Re-classification of Streptomyces venezuelae strains and mining secondary metabolite biosynthetic gene clusters. iScience. 2021 Nov 9;24(12):103410. doi: 10.1016/j.isci.2021.103410. PMID: 34877485; PMCID: PMC8627960.
-

Optimal dimensionality selection for independent component analysis of transcriptomic data
McConn JL, Lamoureux CR, Poudel S, Palsson BO, Sastry AV. Optimal dimensionality selection for independent component analysis of transcriptomic data. BMC Bioinformatics. 2021 Dec 8;22(1):584. doi: 10.1186/s12859-021-04497-7. PMID: 34879815.
-

Escherichia coli Data-Driven Strain Design Using Aggregated Adaptive Laboratory Evolution Mutational Data
Phaneuf PV, Zielinski DC, Yurkovich JT, Johnsen J, Szubin R, Yang L, Kim SH, Schulz S, Wu M, Dalldorf C, Ozdemir E, Lennen RM, Palsson BO, Feist AM. Escherichia coli Data-Driven Strain Design Using Aggregated Adaptive Laboratory Evolution Mutational Data. ACS Synth Biol. 2021 Nov 11. doi: 10.1021/acssynbio.1c00337. Epub ahead of print. PMID: 34762392.
-

proChIPdb: a chromatin immunoprecipitation database for prokaryotic organisms
Decker KT, Gao Y, Rychel K, Al Bulushi T, Chauhan SM, Kim D, Cho BK, Palsson BO. proChIPdb: a chromatin immunoprecipitation database for prokaryotic organisms. Nucleic Acids Res. 2021 Nov 17:gkab1043. doi: 10.1093/nar/gkab1043. Epub ahead of print. PMID: 34791440.
-

Machine Learning Uncovers a Data-Driven Transcriptional Regulatory Network for the Crenarchaeal Thermoacidophile Sulfolobus acidocaldarius
Chauhan Siddharth M., Poudel Saugat, Rychel Kevin, Lamoureux Cameron, Yoo Reo, Al Bulushi Tahani, Yuan Yuan, Palsson Bernhard O., Sastry Anand V. Machine Learning Uncovers a Data-Driven Transcriptional Regulatory Network for the Crenarchaeal Thermoacidophile Sulfolobus acidocaldarius. Frontiers in Microbiology 2021. 12:3213. DOI=10.3389/fmicb.2021.753521
-

RiboRid: A low cost, advanced, and ultra-efficient method to remove ribosomal RNA for bacterial transcriptomics
Choe D, Szubin R, Poudel S, Sastry A, Song Y, Lee Y, Cho S, Palsson B, Cho BK. RiboRid: A low cost, advanced, and ultra-efficient method to remove ribosomal RNA for bacterial transcriptomics. PLoS Genet. 2021 Sep 27;17(9):e1009821. doi: 10.1371/journal.pgen.1009821. Epub ahead of print. PMID: 34570751.
-

Path to improving the life cycle and quality of genome-scale models of metabolism
Seif Y, Palsson BØ. Path to improving the life cycle and quality of genome-scale models of metabolism. Cell Syst. 2021 Sep 22;12(9):842-859. doi: 10.1016/j.cels.2021.06.005. PMID: 34555324; PMCID: PMC8480436.
-

Generation of Pseudomonas putida KT2440 Strains with Efficient Utilization of Xylose and Galactose via Adaptive Laboratory Evolution
Lim HG, Eng T, Banerjee D, Alarcon G, Lau AK, Park M-R, Simmons BA, Palsson BO, Singer SW, Mukhopadhyay A, Feist AM. Generation of Pseudomonas putida KT2440 Strains with Efficient Utilization of Xylose and Galactose via Adaptive Laboratory Evolution. ACS Sustainable Chem Eng [Internet]. American Chemical Society; 2021 Aug 30;9(34):11512–11523. Available from: https://doi.org/10.1021/acssuschemeng.1c03765
-

Machine Learning of Bacterial Transcriptomes Reveals Responses Underlying Differential Antibiotic Susceptibility
Sastry AV, Dillon N, Anand A, Poudel S, Hefner Y, Xu S, Szubin R, Feist AM, Nizet V, Palsson B. Machine Learning of Bacterial Transcriptomes Reveals Responses Underlying Differential Antibiotic Susceptibility. mSphere. 2021 Aug 25;6(4):e0044321. doi: 10.1128/mSphere.00443-21. Epub 2021 Aug 25. PMID: 34431696.
-

Unraveling the functions of uncharacterized transcription factors in Escherichia coli using ChIP-exo
Gao Y, Lim HG, Verkler H, Szubin R, Quach D, Rodionova I, Chen K, Yurkovich JT, Cho BK, Palsson BO. Unraveling the functions of uncharacterized transcription factors in Escherichia coli using ChIP-exo. Nucleic Acids Res. 2021 Aug 24:gkab735. doi: 10.1093/nar/gkab735. Epub ahead of print. PMID: 34428301.
-

Identification of a transcription factor, PunR, that regulates the purine and purine nucleoside transporter punC in E. coli
Rodionova IA, Gao Y, Sastry A, Hefner Y, Lim HG, Rodionov DA, Saier MH Jr, Palsson BO. Identification of a transcription factor, PunR, that regulates the purine and purine nucleoside transporter punC in E. coli. Commun Biol. 2021 Aug 19;4(1):991. doi: 10.1038/s42003-021-02516-0. PMID: 34413462.
-

Genome-scale metabolic modeling reveals key features of a minimal gene set
Lachance JC, Matteau D, Brodeur J, Lloyd CJ, Mih N, King ZA, Knight TF, Feist AM, Monk JM, Palsson BO, Jacques PÉ, Rodrigue S. Genome-scale metabolic modeling reveals key features of a minimal gene set. Mol Syst Biol. 2021 Jul;17(7):e10099. doi: 10.15252/msb.202010099. PMID: 34288418; PMCID: PMC8290834.
-

Computation of condition-dependent proteome allocation reveals variability in the macro and micro nutrient requirements for growth
Lloyd CJ, Monk J, Yang L, Ebrahim A, Palsson BO. Computation of condition-dependent proteome allocation reveals variability in the macro and micro nutrient requirements for growth. PLoS Comput Biol. 2021 Jun 23;17(6):e1007817. doi: 10.1371/journal.pcbi.1007817. PMID: 34161321.
-

Environmental conditions dictate differential evolution of vancomycin resistance in Staphylococcus aureus
Machado H, Seif Y, Sakoulas G, Olson CA, Hefner Y, Anand A, Jones YZ, Szubin R, Palsson BO, Nizet V, Feist AM. Environmental conditions dictate differential evolution of vancomycin resistance in Staphylococcus aureus. Commun Biol. 2021 Jun 25;4(1):793. doi: 10.1038/s42003-021-02339-z. PMID: 34172889.
-

The quantitative metabolome is shaped by abiotic constraints
Akbari A, Yurkovich JT, Zielinski DC, Palsson BO. The quantitative metabolome is shaped by abiotic constraints. Nat Commun. 2021 May 26;12(1):3178. doi: 10.1038/s41467-021-23214-9. PMID: 34039963.
-

Elucidating the Regulatory Elements for Transcription Termination and Posttranscriptional Processing in the Streptomyces clavuligerus Genome.
Hwang S, Lee N, Choe D, Lee Y, Kim W, Jeong Y, Cho S, Palsson BO, Cho BK. Elucidating the Regulatory Elements for Transcription Termination and Posttranscriptional Processing in the Streptomyces clavuligerus Genome. mSystems. 2021 May 4;6(3):e01013-20. doi: 10.1128/mSystems.01013-20. PMID: 33947798.
-

Discovery of novel secondary metabolites encoded in actinomycete genomes through coculture
Kim JH, Lee N, Hwang S, Kim W, Lee Y, Cho S, Palsson BO, Cho BK. Discovery of novel secondary metabolites encoded in actinomycete genomes through coculture. J Ind Microbiol Biotechnol. 2021 Jan 25:kuaa001. doi: 10.1093/jimb/kuaa001. Epub ahead of print. PMID: 33825906.
-

Restoration of fitness lost due to dysregulation of the pyruvate dehydrogenase complex is triggered by ribosomal binding site modifications
Anand A, Olson CA, Sastry AV, Patel A, Szubin R, Yang L, Feist AM, Palsson BO. Restoration of fitness lost due to dysregulation of the pyruvate dehydrogenase complex is triggered by ribosomal binding site modifications. Cell Rep. 2021 Apr 6;35(1):108961. doi: 10.1016/j.celrep.2021.108961. PMID: 33826886.
-

MASSpy: Building, simulating, and visualizing dynamic biological models in Python using mass action kinetics
Haiman ZB, Zielinski DC, Koike Y, Yurkovich JT, Palsson BO. MASSpy: Building, simulating, and visualizing dynamic biological models in Python using mass action kinetics. PLoS Comput Biol. 2021 Jan 28;17(1):e1008208. doi: 10.1371/journal.pcbi.1008208. PMID: 33507922; PMCID: PMC7872247.
-

Experimentally Validated Reconstruction and Analysis of a Genome-Scale Metabolic Model of an Anaerobic Neocallimastigomycota Fungus
Wilken SE, Monk JM, Leggieri PA, Lawson CE, Lankiewicz TS, Seppälä S, Daum CG, Jenkins J, Lipzen AM, Mondo SJ, Barry KW, Grigoriev IV, Henske JK, Theodorou MK, Palsson BO, Petzold LR, O'Malley MA. Experimentally Validated Reconstruction and Analysis of a Genome-Scale Metabolic Model of an Anaerobic Neocallimastigomycota Fungus. mSystems. 2021 Feb 16;6(1):e00002-21. doi: 10.1128/mSystems.00002-21. PMID: 33594000.
-

Independent component analysis recovers consistent regulatory signals from disparate datasets
Sastry AV, Hu A, Heckmann D, Poudel S, Kavvas E, Palsson BO. Independent component analysis recovers consistent regulatory signals from disparate datasets. PLoS Comput Biol. 2021 Feb 2;17(2):e1008647. doi: 10.1371/journal.pcbi.1008647. PMID: 33529205.
-

Pangenome Analytics Reveal Two-Component Systems as Conserved Targets in ESKAPEE Pathogens
Akanksha Rajput, Yara Seif, Kumari Sonal Choudhary, Christopher Dalldorf, Saugat Poudel, Jonathan M. Monk, Bernhard O. Palsson. mSystems Jan 2021, 6 (1) e00981-20; DOI: 10.1128/mSystems.00981-20
-

Bacterial fitness landscapes stratify based on proteome allocation associated with discrete aero-types
Chen K, Anand A, Olson C, Sandberg TE, Gao Y, Mih N, Palsson BO. Bacterial fitness landscapes stratify based on proteome allocation associated with discrete aero-types. PLoS Comput Biol. 2021 Jan 19;17(1):e1008596. doi: 10.1371/journal.pcbi.1008596. Epub ahead of print. PMID: 33465077.
-

Systems and synthetic biology to elucidate secondary metabolite biosynthetic gene clusters encoded in Streptomyces genomes
Lee N, Hwang S, Kim W, Lee Y, Kim JH, Cho S, Kim HU, Yoon YJ, Oh MK, Palsson BO, Cho BK. Systems and synthetic biology to elucidate secondary metabolite biosynthetic gene clusters encoded in Streptomyces genomes. Nat Prod Rep. 2021 Jan 4. doi: 10.1039/d0np00071j. Epub ahead of print. PMID: 33393961.
-

Identifying the effect of vancomycin on health care-associated methicillin-resistant Staphylococcus aureus strains using bacteriological and physiological media
Rajput A, Poudel S, Tsunemoto H, Meehan M, Szubin R, Olson CA, Seif Y, Lamsa A, Dillon N, Vrbanac A, Sugie J, Dahesh S, Monk JM, Dorrestein PC, Knight R, Pogliano J, Nizet V, Feist AM, Palsson BO. Identifying the effect of vancomycin on health care-associated methicillin-resistant Staphylococcus aureus strains using bacteriological and physiological media. Gigascience. 2021 Jan 9;10(1):giaa156. doi: 10.1093/gigascience/giaa156. PMID: 33420779.
-

DeepTFactor: A deep learning-based tool for the prediction of transcription factors
DeepTFactor: A deep learning-based tool for the prediction of transcription factors. Gi Bae Kim, Ye Gao, Bernhard O. Palsson, Sang Yup Lee. Proceedings of the National Academy of Sciences Jan 2021, 118 (2) e2021171118; DOI:10.1073/pnas.2021171118
-

The Expanding Computational Toolbox for Engineering Microbial Phenotypes at the Genome Scale
Zielinski, D.C.; Patel, A.; Palsson, B.O. The Expanding Computational Toolbox for Engineering Microbial Phenotypes at the Genome Scale. Microorganisms 2020, 8, 2050.
-

Machine learning uncovers independently regulated modules in the Bacillus subtilis transcriptome
Rychel, K., Sastry, A.V. & Palsson, B.O. Machine learning uncovers independently regulated modules in the Bacillus subtilis transcriptome. Nat Commun 11, 6338 (2020). https://doi.org/10.1038/s41467-020-20153-9
-

High-Quality Genome-Scale Models From Error-Prone, Long-Read Assemblies
Broddrick Jared T., Szubin Richard, Norsigian Charles J., Monk Jonathan M., Palsson Bernhard O., Parenteau Mary N., High-Quality Genome-Scale Models From Error-Prone, Long-Read Assemblies, Frontiers in Microbiology 11:2829 2020. DOI:10.3389/fmicb.2020.596626
-

Scalable computation of intracellular metabolite concentrations
Amir Akbari, Bernhard O. Palsson, Scalable computation of intracellular metabolite concentrations, Computers & Chemical Engineering, 2020, 107164, ISSN 0098-1354, https://doi.org/10.1016/j.compchemeng.2020.107164 (http://www.sciencedirect.com/science/article/pii/S0098135420308619)
-

Elucidation of Regulatory Modes for Five Two-Component Systems in Escherichia coli Reveals Novel Relationships
Choudhary KS, Kleinmanns JA, Decker K, Sastry AV, Gao Y, Szubin R, Seif Y, Palsson BO. Elucidation of Regulatory Modes for Five Two-Component Systems in Escherichia coli Reveals Novel Relationships. mSystems. 2020 Nov 10;5(6):e00980-20. doi: 10.1128/mSystems.00980-20. PMID: 33172971.
-

iModulonDB: a knowledgebase of microbial transcriptional regulation derived from machine learning
Rychel K, Decker K, Sastry AV, Phaneuf PV, Poudel S, Palsson BO. iModulonDB: a knowledgebase of microbial transcriptional regulation derived from machine learning. Nucleic Acids Res. 2020 Oct 12:gkaa810. doi: 10.1093/nar/gkaa810. Epub ahead of print. PMID: 33045728.
-

Systems biology analysis of the Clostridioides difficile core-genome contextualizes microenvironmental evolutionary pressures leading to genotypic and phenotypic divergence
Norsigian, C.J., Danhof, H.A., Brand, C.K. et al. Systems biology analysis of the Clostridioides difficile core-genome contextualizes microenvironmental evolutionary pressures leading to genotypic and phenotypic divergence. npj Syst Biol Appl 6, 31 (2020). https://doi.org/10.1038/s41540-020-00151-9
-

Genome-scale metabolic models highlight stage-specific differences in essential metabolic pathways in Trypanosoma cruzi
Shiratsubaki IS, Fang X, Souza ROO, Palsson BO, Silber AM, Siqueira-Neto JL. Genome-scale metabolic models highlight stage-specific differences in essential metabolic pathways in Trypanosoma cruzi. PLoS Negl Trop Dis. 2020 Oct 6;14(10):e0008728. doi: 10.1371/journal.pntd.0008728. Epub ahead of print. PMID: 33021977.
-

Gastrointestinal Surgery for Inflammatory Bowel Disease Persistently Lowers Microbiome and Metabolome Diversity
Fang X, Vázquez-Baeza Y, Elijah E, Vargas F, Ackermann G, Humphrey G, Lau R, Weldon KC, Sanders JG, Panitchpakdi M, Carpenter C, Jarmusch AK, Neill J, Miralles A, Dulai P, Singh S, Tsai M, Swafford AD, Smarr L, Boyle DL, Palsson BO, Chang JT, Dorrestein PC, Sandborn WJ, Knight R, Boland BS. Gastrointestinal Surgery for Inflammatory Bowel Disease Persistently Lowers Microbiome and Metabolome Diversity. Inflamm Bowel Dis. 2020 Oct 7:izaa262. doi: 10.1093/ibd/izaa262. Epub ahead of print. PMID: 33026068.
-

Generation of ionic liquid tolerant Pseudomonas putida KT2440 strains via adaptive laboratory evolution
Lim, HG ; Fong, B ; Alarcon, G ; Magurudeniya, HD ; Eng, T ; Szubin, R; Olson, CA ; Palsson, BO; Gladden, JM ; Simmons, BA ; Mukhopadhyay, A Singer, SW ; Feist, AM. Generation of ionic liquid tolerant Pseudomonas putida KT2440 strains via adaptive laboratory evolution. GREEN CHEMISTRY 22:5677-5690.
-

The Bitome: digitized genomic features reveal fundamental genome organization
Lamoureux CR, Choudhary KS, King ZA, Sandberg TE, Gao Y, Sastry AV, Phaneuf PV, Choe D, Cho BK, Palsson BO. The Bitome: digitized genomic features reveal fundamental genome organization. Nucleic Acids Res. 2020 Sep 25:gkaa774. doi: 10.1093/nar/gkaa774. Epub ahead of print. PMID: 32976587.
-

Reconstructing organisms in silico: genome-scale models and their emerging applications
Fang X, Lloyd CJ, Palsson BO. Reconstructing organisms in silico: genome-scale models and their emerging applications. Nat Rev Microbiol. 2020 Sep 21. doi: 10.1038/s41579-020-00440-4. Epub ahead of print. PMID: 32958892.
-

Kinetic profiling of metabolic specialists demonstrates stability and consistency of in vivo enzyme turnover numbers
Heckmann D, Campeau A, Lloyd CJ, et al. Kinetic profiling of metabolic specialists demonstrates stability and consistency of in vivo enzyme turnover numbers [published online ahead of print, 2020 Sep 1]. Proc Natl Acad Sci U S A. 2020;202001562. doi:10.1073/pnas.2001562117
-

SBML Level 3: an extensible format for the exchange and reuse of biological models
Keating SM, Waltemath D, König M, et al. SBML Level 3: an extensible format for the exchange and reuse of biological models. Mol Syst Biol. 2020;16(8):e9110. doi:10.15252/msb.20199110
-

System-level understanding of gene expression and regulation for engineering secondary metabolite production in Streptomyces
Lee Y, Lee N, Hwang S, et al. System-level understanding of gene expression and regulation for engineering secondary metabolite production in Streptomyces [published online ahead of print, 2020 Aug 10]. J Ind Microbiol Biotechnol. 2020;10.1007/s10295-020-02298-0. doi:10.1007/s10295-020-02298-0
-

Synthetic cross-phyla gene replacement and evolutionary assimilation of major enzymes
Sandberg TE, Szubin R, Phaneuf PV, Palsson BO. Synthetic cross-phyla gene replacement and evolutionary assimilation of major enzymes [published online ahead of print, 2020 Aug 10]. Nat Ecol Evol. 2020;10.1038/s41559-020-1271-x. doi:10.1038/s41559-020-1271-x
-

Causal mutations from adaptive laboratory evolution are outlined by multiple scales of genome annotations and condition-specificity
Phaneuf PV, Yurkovich JT, Heckmann D, et al. Causal mutations from adaptive laboratory evolution are outlined by multiple scales of genome annotations and condition-specificity. BMC Genomics. 2020;21(1):514. Published 2020 Jul 25. doi:10.1186/s12864-020-06920-4
-

Impact of insertion sequences on convergent evolution of Shigella species
Hawkey J, Monk JM, Billman-Jacobe H, Palsson B, Holt KE. Impact of insertion sequences on convergent evolution of Shigella species. PLoS Genet. 2020;16(7):e1008931. Published 2020 Jul 9. doi:10.1371/journal.pgen.1008931
-

Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces
Lee N, Hwang S, Kim J, Cho S, Palsson B, Cho BK. Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput Struct Biotechnol J. 2020;18:1548-1556. Published 2020 Jun 21. doi:10.1016/j.csbj.2020.06.024
-

Independent component analysis of E. coli's transcriptome reveals thecellular processes that respond to heterologous gene expression
Tan J, Sastry AV, Fremming KS, et al. Independent component analysis of E. coli's transcriptome reveals the cellular processes that respond to heterologous gene expression [published online ahead of print, 2020 Jul 22]. Metab Eng. 2020;S1096-7176(20)30111-7. doi:10.1016/j.ymben.2020.07.002
-

Redefining fundamental concepts of transcription initiation in bacteria
Mejía-Almonte C, Busby SJW, Wade JT, et al. Redefining fundamental concepts of transcription initiation in bacteria [published online ahead of print, 2020 Jul 14]. Nat Rev Genet. 2020;10.1038/s41576-020-0254-8. doi:10.1038/s41576-020-0254-8
-

Synthesizing Systems Biology Knowledge from Omics Using Genome-Scale Models
Dahal S, Yurkovich JT, Xu H, Palsson BO, Yang L. Synthesizing Systems Biology Knowledge from Omics Using Genome-Scale Models [published online ahead of print, 2020 Jun 24]. Proteomics. 2020;e1900282. doi:10.1002/pmic.201900282
-

Revealing 29 sets of independently modulated genes in Staphylococcus aureus, their regulators, and role in key physiological response
Poudel S, Tsunemoto H, Seif Y, et al. Revealing 29 sets of independently modulated genes in Staphylococcus aureus, their regulators, and role in key physiological response. Proc Natl Acad Sci U S A. 2020;202008413. doi:10.1073/pnas.2008413117
-

Comparative Genomics Determines Strain-Dependent Secondary Metabolite Production in Streptomyces venezuelae Strains
Kim W, Lee N, Hwang S, et al. Comparative Genomics Determines Strain-Dependent Secondary Metabolite Production in Streptomyces venezuelae Strains. Biomolecules. 2020;10(6):E864. Published 2020 Jun 5. doi:10.3390/biom10060864
-

Microbial Synthesis of Human-Hormone Melatonin at Gram Scales
Luo H, Schneider K, Christensen U, Lei Y, Herrgard M, Palsson BØ. Microbial Synthesis of Human-Hormone Melatonin at Gram Scales [published online ahead of print, 2020 Jun 5]. ACS Synth Biol. 2020;10.1021/acssynbio.0c00065. doi:10.1021/acssynbio.0c00065
-

Adaptations of Escherichia Coli Strains to Oxidative Stress Are Reflected in Properties of Their Structural Proteomes
Mih N, Monk JM, Fang X, et al. Adaptations of Escherichia coli strains to oxidative stress are reflected in properties of their structural proteomes. BMC Bioinformatics. 2020;21(1):162. Published 2020 Apr 29. doi:10.1186/s12859-020-3505-y
-

Multiplex secretome engineering enhances recombinant protein production and purity
Kol S, Ley D, Wulff T, et al. Multiplex secretome engineering enhances recombinant protein production and purity. Nat Commun. 2020;11(1):1908. Published 2020 Apr 20. doi:10.1038/s41467-020-15866-w
-

Repurposing Modular Polyketide Synthases and Non-ribosomal Peptide Synthetases for Novel Chemical Biosynthesis
Hwang S, Lee N, Cho S, Palsson B, Cho BK. Repurposing Modular Polyketide Synthases and Non-ribosomal Peptide Synthetases for Novel Chemical Biosynthesis. Front. Mol. Biosci., 15 May 2020 https://doi.org/10.3389/fmolb.2020.00087
-

Transcriptome and translatome profiles of Streptomyces species in different growth phases.
Transcriptome and translatome profiles of Streptomyces species in different growth phases. Kim W, Hwang S, Lee N, Lee Y, Cho S, Palsson B, Cho BK. Sci Data. 2020 May 8;7(1):138. doi: 10.1038/s41597-020-0476-9. PMID: 32385251
-

A biochemically-interpretable machine learning classifier for microbial GWAS
Kavvas ES, Yang L, Monk JM, Heckmann D, Palsson BO. A biochemically-interpretable machine learning classifier for microbial GWAS. Nat Commun. 2020;11(1):2580. Published 2020 May 22. doi:10.1038/s41467-020-16310-9
-

Genome Sequence Comparison of Staphylococcus aureus TX0117 and a Beta-Lactamase-Cured Derivative Shows Increased Cationic Peptide Resistance Accompanying Mutations in relA and mnaA
Sales MJ, Sakoulas G, Szubin R, Palsson B, Arias C, Singh KV, Murray BE, Monk JM. Genome Sequence Comparison of Staphylococcus aureus TX0117 and a Beta-Lactamase-Cured Derivative Shows Increased Cationic Peptide Resistance Accompanying Mutations in relA and mnaA. Microbiol Resour Announc. 2020 Apr 30;9(18). pii: e01515-19. doi: 10.1128/MRA.01515-19. PubMed PMID: 32354985.
-

Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis
Minasov G, Lam MR, Rosas Lemus M, Sławek J, Woinska M, Shabalin IG, Shuvalova L, Palsson BØ, Godzik A, Minor W, Satchell KJF. Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis. Protein Sci. 2020 Apr 19. doi: 10.1002/pro.3876. [Epub ahead of print] PubMed PMID: 32306515.
-

Reconstruction and Validation of a Genome-Scale Metabolic Model of Streptococcus oralis (iCJ415), a Human Commensal and Opportunistic Pathogen
Jensen CS, Norsigian CJ, Fang X, Nielsen XC, Christensen JJ, Palsson BO, Monk JM. Reconstruction and Validation of a Genome-Scale Metabolic Model of Streptococcus oralis (iCJ415), a Human Commensal and Opportunistic Pathogen. Front Genet. 2020 Mar 3;11:116. doi: 10.3389/fgene.2020.00116. eCollection 2020. PubMed PMID: 32194617; PubMed Central PMCID: PMC7063969.
-

An atlas of human metabolism
Robinson JL, Kocabaş P, Wang H, Cholley PE, Cook D, Nilsson A, Anton M, Ferreira R, Domenzain I, Billa V, Limeta A, Hedin A, Gustafsson J, Kerkhoven EJ, Svensson LT, Palsson BO, Mardinoglu A, Hansson L, Uhlén M, Nielsen J. An atlas of human metabolism. Sci Signal. 2020 Mar 24;13(624). pii: eaaz1482. doi: 10.1126/scisignal.aaz1482. PubMed PMID: 32209698.
-

STATR: A simple analysis pipeline of Ribo-Seq in bacteria
Choe D, Palsson B, Cho BK. STATR: A simple analysis pipeline of Ribo-Seq in bacteria. J Microbiol. 2020 Mar;58(3):217-226. doi: 10.1007/s12275-020-9536-2. Epub 2020 Jan 28. PubMed PMID: 31989542.
-

Genetic Determinants Enabling Medium-Dependent Adaptation to Nafcillin in Methicillin-Resistant Staphylococcus aureus
Salazar MJ, Machado H, Dillon NA, Tsunemoto H, Szubin R, Dahesh S, Pogliano J, Sakoulas G, Palsson BO, Nizet V, Feist AM. Genetic Determinants Enabling Medium-Dependent Adaptation to Nafcillin in Methicillin-Resistant Staphylococcus aureus. mSystems. 2020 Mar 31;5(2). pii: e00828-19. doi: 10.1128/mSystems.00828-19. PubMed PMID: 32234776.
-

Directed Metabolic Pathway Evolution Enables Functional Pterin-Dependent Aromatic-Amino-Acid Hydroxylation in Escherichia coli
Luo H, Yang L, Kim SH, Wulff T, Feist AM, Herrgard M, Palsson BØ. Directed Metabolic Pathway Evolution Enables Functional Pterin-Dependent Aromatic-Amino-Acid Hydroxylation in Escherichia coli. ACS Synth Biol. 2020 Mar 20;9(3):494-499. doi: 10.1021/acssynbio.9b00488. Epub 2020 Mar 9. PubMed PMID: 32149495.
-

Genome-scale reconstructions of the mammalian secretory pathway predict metabolic costs and limitations of protein secretion
Gutierrez JM, Feizi A, Li S, Kallehauge TB, Hefzi H, Grav LM, Ley D, Baycin Hizal D, Betenbaugh MJ, Voldborg B, Faustrup Kildegaard H, Min Lee G, Palsson BO, Nielsen J, Lewis NE. Genome-scale reconstructions of the mammalian secretory pathway predict metabolic costs and limitations of protein secretion. Nat Commun. 2020 Jan 2;11(1):68. doi: 10.1038/s41467-019-13867-y. PubMed PMID: 31896772; PubMed Central PMCID: PMC6940358.
-

MEMOTE for standardized genome-scale metabolic model testing
Lieven C, Beber ME, Olivier BG, Bergmann FT, Ataman M, Babaei P, Bartell JA, Blank LM, Chauhan S, Correia K, Diener C, Dräger A, Ebert BE, Edirisinghe JN, Faria JP, Feist AM, Fengos G, Fleming RMT, García-Jiménez B, Hatzimanikatis V, van Helvoirt W, Henry CS, Hermjakob H, Herrgård MJ, Kaafarani A, Kim HU, King Z, Klamt S, Klipp E, Koehorst JJ, König M, Lakshmanan M, Lee DY, Lee SY, Lee S, Lewis NE, Liu F, Ma H, Machado D, Mahadevan R, Maia P, Mardinoglu A, Medlock GL, Monk JM, Nielsen J, Nielsen LK, Nogales J, Nookaew I, Palsson BO, Papin JA, Patil KR, Poolman M, Price ND, Resendis-Antonio O, Richelle A, Rocha I, Sánchez BJ, Schaap PJ, Malik Sheriff RS, Shoaie S, Sonnenschein N, Teusink B, Vilaça P, Vik JO, Wodke JAH, Xavier JC, Yuan Q, Zakhartsev M, Zhang C. MEMOTE for standardized genome-scale metabolic model testing. Nat Biotechnol. 2020 Mar 2. doi:10.1038/s41587-020-0446-y. [Epub ahead of print] PubMed PMID: 32123384.
-

Metabolic and genetic basis for auxotrophies in Gram-negative species
Seif Y, Choudhary KS, Hefner Y, Anand A, Yang L, Palsson BO. Metabolic and genetic basis for auxotrophies in Gram-negative species. Proc Natl Acad Sci U S A. 2020 Mar 4. pii: 201910499. doi: 10.1073/pnas.1910499117. [Epub ahead of print] PubMed PMID: 32132208.
-

Machine learning with random subspace ensembles identifies antimicrobial resistance determinants from pan-genomes of three pathogens
Hyun JC, Kavvas ES, Monk JM, Palsson BO. Machine learning with random subspace ensembles identifies antimicrobial resistance determinants from pan-genomes of three pathogens. PLoS Comput Biol. 2020 Mar 2;16(3):e1007608. doi: 10.1371/journal.pcbi.1007608. [Epub ahead of print] PubMed PMID: 32119670.
-

Thirty complete Streptomyces genome sequences for mining novel secondary metabolite biosynthetic gene clusters
Lee N, Kim W, Hwang S, Lee Y, Cho S, Palsson B, Cho BK. Thirty complete Streptomyces genome sequences for mining novel secondary metabolite biosynthetic gene clusters. Sci Data. 2020 Feb 13;7(1):55. doi: 10.1038/s41597-020-0395-9. PubMed PMID: 32054853.
-

Iron competition triggers antibiotic biosynthesis in Streptomyces coelicolor during coculture with Myxococcus xanthus
Lee N, Kim W, Chung J, Lee Y, Cho S, Jang KS, Kim SC, Palsson B, Cho BK. Iron competition triggers antibiotic biosynthesis in Streptomyces coelicolor during coculture with Myxococcus xanthus. ISME J. 2020 Jan 28. doi:10.1038/s41396-020-0594-6. [Epub ahead of print] PubMed PMID: 31992858.
-

A workflow for generating multi-strain genome-scale metabolic models of prokaryotes
Norsigian CJ, Fang X, Seif Y, Monk JM, Palsson BO. A workflow for generating multi-strain genome-scale metabolic models of prokaryotes. Nat Protoc. 2020 Jan;15(1):1-14. doi: 10.1038/s41596-019-0254-3. Epub 2019 Dec 20. PubMed PMID: 31863076.
-

Systems-level analysis of NalD mutation, a recurrent driver of rapid drug resistance in acute Pseudomonas aeruginosa infection
Yan J, Estanbouli H, Liao C, Kim W, Monk JM, Rahman R, Kamboj M, Palsson BO, Qiu W, Xavier JB. Systems-level analysis of NalD mutation, a recurrent driver of rapid drug resistance in acute Pseudomonas aeruginosa infection. PLoS Comput Biol. 2019 Dec 20;15(12):e1007562. doi: 10.1371/journal.pcbi.1007562. [Epub ahead of print] PubMed PMID: 31860667.
-

Profiling the effect of nafcillin on HA-MRSA D712 using bacteriological and physiological media
Rajput A, Poudel S, Tsunemoto H, Meehan M, Szubin R, Olson CA, Lamsa A, Seif Y, Dillon N, Vrbanac A, Sugie J, Dahesh S, Monk JM, Dorrestein PC, Knight R, Nizet V, Palsson BO, Feist AM, Pogliano J. Profiling the effect of nafcillin on HA-MRSA D712 using bacteriological and physiological media. Sci Data. 2019 Dec 17;6(1):322. doi: 10.1038/s41597-019-0331-z. PubMed PMID: 31848353.
-

Structure of galactarate dehydratase, a new fold in an enolase involved in bacterial fitness after antibiotic treatment
Rosas-Lemus M, Minasov G, Shuvalova L, Wawrzak Z, Kiryukhina O, Mih N, Jaroszewski L, Palsson B, Godzik A, Satchell KJF. Structure of galactarate dehydratase, a new fold in an enolase involved in bacterial fitness after antibiotic treatment. Protein Sci. 2019 Dec 6. doi: 10.1002/pro.3796. [Epub ahead of print] PubMed PMID: 31811683.
-

Minimal cells, maximal knowledge
Lachance JC, Rodrigue S, Palsson BO. Minimal cells, maximal knowledge. Elife. 2019 Mar 12;8. pii: e45379. doi: 10.7554/eLife.45379. PubMed PMID: 30860025; PubMed Central PMCID: PMC6414198.
-

BiGG Models 2020: multi-strain genome-scale models and expansion across the phylogenetic tree
Norsigian CJ, Pusarla N, McConn JL, Yurkovich JT, Dräger A, Palsson BO, King Z. BiGG Models 2020: multi-strain genome-scale models and expansion across the phylogenetic tree. Nucleic Acids Res. 2019 Nov 7. pii: gkz1054. doi: 10.1093/nar/gkz1054. [Epub ahead of print] PubMed PMID: 31696234.
-

Genome-scale model of metabolism and gene expression provides a multi-scale description of acid stress responses in Escherichia coli
Du B, Yang L, Lloyd CJ, Fang X, Palsson BO. Genome-scale model of metabolism and gene expression provides a multi-scale description of acid stress responses in Escherichia coli. PLoS Comput Biol. 2019 Dec 6;15(12):e1007525. doi: 10.1371/journal.pcbi.1007525. eCollection 2019 Dec. PubMed PMID: 31809503.
-

The Escherichia coli transcriptome mostly consists of independently regulated modules
Sastry AV, Gao Y, Szubin R, Hefner Y, Xu S, Kim D, Choudhary KS, Yang L, King ZA, Palsson BO. The Escherichia coli transcriptome mostly consists of independently regulated modules. Nat Commun. 2019 Dec 4;10(1):5536. doi: 10.1038/s41467-019-13483-w. PubMed PMID: 31797920.
-

Expanding the uses of genome‐scale models with protein structures
Mih N, Palsson BO. Expanding the uses of genome-scale models with protein structures. Mol Syst Biol. 2019 Nov;15(11):e8601. doi: 10.15252/msb.20188601. PubMed PMID: 31777175; PubMed Central PMCID: PMC6880182.
-

Adaptive evolution reveals a tradeoff between growth rate and oxidative stress during naphthoquinone-based aerobic respiration
Proc Natl Acad Sci U S A. 2019 Nov 25. pii: 201909987. doi: 10.1073/pnas.1909987116. [Epub ahead of print] PubMed PMID: 31767748.
-

High-quality genome-scale metabolic modeling of Pseudomonas putida highlights its broad metabolic capabilities
Nogales J, Mueller J, Gudmundsson S, Canalejo FJ, Duque E, Monk J, Feist AM, Ramos JL, Niu W, Palsson BO. High-quality genome-scale metabolic modeling of Pseudomonas putida highlights its broad metabolic capabilities. Environ Microbiol. 2019 Oct 27. doi: 10.1111/1462-2920.14843. [Epub ahead of print] PubMed PMID: 31657101.
-

OxyR is a convergent target for mutations acquired during adaptation to oxidative stress-prone metabolic states
Anand A, Chen K, Catoiu E, Sastry AV, Olson CA, Sandberg TE, Seif Y, Xu S, Szubin R, Yang L, Feist AM, Palsson BO. OxyR is a convergent target for mutations acquired during adaptation to oxidative stress-prone metabolic states. Mol Biol Evol. 2019 Oct 25. pii: msz251. doi: 10.1093/molbev/msz251. [Epub ahead of print] PubMed PMID: 31651953.
-

Adaptive laboratory evolution of Escherichia coli under acid stress
Du et al., Microbiology DOI 10.1099/mic.0.000867 (2019)
-

Evolution and regulation of nitrogen flux through compartmentalized metabolic networks in a marine diatom
Smith SR, Dupont CL, McCarthy JK, Broddrick JT, Oborník M, Horák A, Füssy Z, Cihlář J, Kleessen S, Zheng H, McCrow JP, Hixson KK, Araújo WL, Nunes-Nesi A, Fernie A, Nikoloski Z, Palsson BO, Allen AE. Evolution and regulation of nitrogen flux through compartmentalized metabolic networks in a marine diatom. Nat Commun. 2019 Oct 7;10(1):4552. doi: 10.1038/s41467-019-12407-y. PubMed PMID: 31591397.
-

The genetic basis for adaptation of model-designed syntrophic co-cultures
Lloyd CJ, King ZA, Sandberg TE, Hefner Y, Olson CA, Phaneuf PV, O'Brien EJ, Sanders JG, Salido RA, Sanders K, Brennan C, Humphrey G, Knight R, Feist AM. The genetic basis for adaptation of model-designed syntrophic co-cultures. PLoS Comput Biol. 2019 Mar 1;15(3):e1006213. doi: 10.1371/journal.pcbi.1006213. eCollection 2019 Mar. PubMed PMID: 30822347; PubMed Central PMCID: PMC6415869.
-

The Transcription Unit Architecture of Streptomyces lividans TK24
Lee Y, Lee N, Jeong Y, Hwang S, Kim W, Cho S, Palsson BO, Cho BK. The Transcription Unit Architecture of Streptomyces lividans TK24. Frontiers in Microbiology 10:2074. 2019.
-

Inactivation of a Mismatch-Repair System Diversifies Genotypic Landscape of Escherichia coli During Adaptive Laboratory Evolution
Kang M, Kim K, Choe D, Cho S, Kim SC, Palsson B, Cho BK. Inactivation of a Mismatch-Repair System Diversifies Genotypic Landscape of Escherichia coli During Adaptive Laboratory Evolution. Front Microbiol. 2019 Aug 16;10:1845. doi: 10.3389/fmicb.2019.01845. eCollection 2019. PubMed PMID: 31474949; PubMed Central PMCID: PMC6706779.
-

A defined minimal medium for systems analyses of Staphylococcus aureus reveals strain-specific metabolic requirements
Machado H, Weng LL, Dillon N, Seif Y, Holland M, Pekar JE, Monk JM, Nizet V, Palsson BO, Feist AM. A defined minimal medium for systems analyses of Staphylococcus aureus reveals strain-specific metabolic requirements. Appl Environ Microbiol. 2019 Aug 30. pii: AEM.01773-19. doi: 10.1128/AEM.01773-19. [Epub ahead of print] PubMed PMID: 31471305.
-

Systems Biology and Pangenome of Salmonella O-Antigens
Seif Y, Monk JM, Machado H, Kavvas E, Palsson BO. Systems Biology and Pangenome of Salmonella O-Antigens. mBio. 2019 Aug 27;10(4). pii: e01247-19. doi: 10.1128/mBio.01247-19. PubMed PMID: 31455646; PubMed Central PMCID: PMC6712391.
-

The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology
Sandberg TE, Salazar MJ, Weng LL, Palsson BO, Feist AM. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab Eng. 2019 Aug 8. pii: S1096-7176(19)30153-3. doi: 10.1016/j.ymben.2019.08.004. [Epub ahead of print] Review. PubMed PMID: 31401242.
-

Comparative Genome-Scale Metabolic Modeling of Metallo-Beta-Lactamase-Producing Multidrug-Resistant Klebsiella pneumoniae Clinical Isolates
Norsigian CJ, Attia H, Szubin R, Yassin AS, Palsson BØ, Aziz RK, Monk JM. Comparative Genome-Scale Metabolic Modeling of Metallo-Beta-Lactamase-Producing Multidrug-Resistant Klebsiella pneumoniae Clinical Isolates. Front Cell Infect Microbiol. 2019 May 24;9:161. doi: 10.3389/fcimb.2019.00161. eCollection 2019. PubMed PMID: 31179245; PubMed Central PMCID: PMC6543805.
-

Metabolic Systems Analysis of Shock-Induced Endotheliopathy (SHINE) in Trauma: A New Research Paradigm
Henriksen HH, McGarrity S, SigurÐardóttir RS, Nemkov T, D'Alessandro A, Palsson BO, Stensballe J, Wade CE, Rolfsson Ó, Johansson PI. Metabolic Systems Analysis of Shock-Induced Endotheliopathy (SHINE) in Trauma: A New Research Paradigm. Ann Surg. 2019 Jun 26. doi: 10.1097/SLA.0000000000003307. [Epub ahead of print] PubMed PMID: 31274658.
-

Cellular responses to reactive oxygen species are predicted from molecular mechanisms
Yang L, Mih N, Anand A, Park JH, Tan J, Yurkovich JT, Monk JM, Lloyd CJ, Sandberg TE, Seo SW, Kim D, Sastry AV, Phaneuf P, Gao Y, Broddrick JT, Chen K, Heckmann D, Szubin R, Hefner Y, Feist AM, Palsson BO. Cellular responses to reactive oxygen species are predicted from molecular mechanisms. Proc Natl Acad Sci U S A. 2019 Jul 3. pii: 201905039. doi: 10.1073/pnas.1905039116. [Epub ahead of print] PubMed PMID: 31270234.
-

Synthetic Biology Tools for Novel Secondary Metabolite Discovery in Streptomyces
Lee N, Hwang S, Lee Y, Cho S, Palsson B, Cho BK. Synthetic Biology Tools for Novel Secondary Metabolite Discovery in Streptomyces. J Microbiol Biotechnol. 2019 May 28;29(5):667-686. doi: 10.4014/jmb.1904.04015. Review. PubMed PMID: 31091862.
-

Laboratory evolution reveals a two-dimensional rate-yield tradeoff in microbial metabolism
Cheng C, O'Brien EJ, McCloskey D, Utrilla J, Olson C, LaCroix RA, Sandberg TE, Feist AM, Palsson BO, King ZA. Laboratory evolution reveals a two-dimensional rate-yield tradeoff in microbial metabolism. PLoS Comput Biol. 2019 Jun 3;15(6):e1007066. doi: 10.1371/journal.pcbi.1007066. [Epub ahead of print] PubMed PMID: 31158228.
-

Primary transcriptome and translatome analysis determines transcriptional and translational regulatory elements encoded in the Streptomyces clavuligerus genome
Hwang S, Lee N, Jeong Y, Lee Y, Kim W, Cho S, Palsson BO, Cho BK. Primary transcriptome and translatome analysis determines transcriptional and translational regulatory elements encoded in the Streptomyces clavuligerus genome. Nucleic Acids Res. 2019 May 27. pii: gkz471. doi: 10.1093/nar/gkz471. [Epub ahead of print] PubMed PMID: 31131406.
-

BOFdata: Generating biomass objective functions for genome-scale metabolic models from experimental data
Lachance JC, Lloyd CJ, Monk JM, Yang L, Sastry AV, Seif Y, Palsson BO, Rodrigue S, Feist AM, King ZA, Jacques PÉ. BOFdat: Generating biomass objective functions for genome-scale metabolic models from experimental data. PLoS Comput Biol. 2019 Apr 22;15(4):e1006971. doi: 10.1371/journal.pcbi.1006971. eCollection 2019 Apr. PubMed PMID: 31009451.
-

Characterization of CA-MRSA TCH1516 exposed to nafcillin in bacteriological and physiological media
Poudel S, Tsunemoto H, Meehan M, Szubin R, Olson CA, Lamsa A, Seif Y, Dillon N, Vrbanac A, Sugie J, Dahesh S, Monk JM, Dorrestein PC, Pogliano J, Knight R, Nizet V, Palsson BO, Feist AM. Characterization of CA-MRSA TCH1516 exposed to nafcillin in bacteriological and physiological media. Sci Data. 2019 Apr 26;6(1):43. doi: 10.1038/s41597-019-0051-4. PubMed PMID: 31028276; PubMed Central PMCID: PMC6486602.
-

A White-Box Machine Learning Approach for Revealing Antibiotic Mechanisms of Action
Yang et al., 2019, Cell 177, 1–13 May 30, 2019
-

Cross-compartment metabolic coupling enables flexible photoprotective mechanisms in the diatom Phaeodactylum tricornutum
Broddrick JT, Du N, Smith SR, Tsuji Y, Jallet D, Ware MA, Peers G, Matsuda Y, Dupont CL, Mitchell BG, Palsson BO, Allen AE. Cross-compartment metabolic coupling enables flexible photoprotective mechanisms in the diatom Phaeodactylum tricornutum. New Phytol. 2019 May;222(3):1364-1379. doi: 10.1111/nph.15685. Epub 2019 Feb 14. PubMed PMID: 30636322.
-

Enzyme promiscuity shapes adaptation to novel growth substrates
Guzmán GI, Sandberg TE, LaCroix RA, Nyerges Á, Papp H, de Raad M, King ZA, Hefner Y, Northen TR, Notebaart RA, Pál C, Palsson BO, Papp B, Feist AM. Enzyme promiscuity shapes adaptation to novel growth substrates. Mol Syst Biol. 2019 Apr 8;15(4):e8462. doi: 10.15252/msb.20188462. PubMed PMID: 30962359; PubMed Central PMCID: PMC6452873.
-

Coupling S-adenosylmethionine-dependent methylation to growth: Design and uses
Luo H, Hansen ASL, Yang L, Schneider K, Kristensen M, Christensen U, Christensen HB, Du B, Özdemir E, Feist AM, Keasling JD, Jensen MK, Herrgård MJ, Palsson BO. Coupling S-adenosylmethionine-dependent methylation to growth: Design and uses. PLoS Biol. 2019 Mar 11;17(3):e2007050. doi: 10.1371/journal.pbio.2007050. eCollection 2019 Mar. PubMed PMID: 30856169; PubMed Central PMCID: PMC6411097.
-

DynamicME: dynamic simulation and refinement of integrated models of metabolism and protein expression
Yang L, Ebrahim A, Lloyd CJ, Saunders MA, Palsson BO. DynamicME: dynamic simulation and refinement of integrated models of metabolism and protein expression. BMC Syst Biol. 2019 Jan 9;13(1):2. doi: 10.1186/s12918-018-0675-6. PubMed PMID: 30626386; PubMed Central PMCID: PMC6327497.
-

The y-ome defines the 35% of Escherichia coli genes that lack experimental evidence of function
Ghatak S, King ZA, Sastry A, Palsson BO. 2019. The y-ome defines the 35% of Escherichia coli genes that lack experimental evidence of function. Nucleic Acids Research.
-

Pseudogene repair driven by selection pressure applied in experimental evolution.
Anand A, Olson CA, Yang L, Sastry AV, Catoiu E, Choudhary KSonal, Phaneuf PV, Sandberg TE, Xu S, Hefner Y et al.. 2019. Pseudogene repair driven by selection pressure applied in experimental evolution.. Nat Microbiol.
-

Adaptive laboratory evolution of a genome-reduced Escherichia coli
Choe D, Lee JHyoung, Yoo M, Hwang S, Sung BHyun, Cho S, Palsson B, Kim SChang, Cho B-K. 2019. Adaptive laboratory evolution of a genome-reduced Escherichia coli. Nature Communications.
-

Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0.
Heirendt L, Arreckx S, Pfau T, Mendoza SN, Richelle A, Heinken A, Haraldsdóttir HS, Wachowiak J, Keating SM, Vlasov V et al.. 2019. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0.. Nat Protoc.
-

A computational knowledge-base elucidates the response of Staphylococcus aureus to different media types.
Seif Y, Monk JM, Mih N, Tsunemoto H, Poudel S, Zuniga C, Broddrick J, Zengler K, Palsson BO. 2019. A computational knowledge-base elucidates the response of Staphylococcus aureus to different media types.. PLoS Comput Biol. 15(1):e1006644.
-

Escher-FBA: a web application for interactive flux balance analysis
Rowe E, Palsson BO, King ZA. 2018. Escher-FBA: a web application for interactive flux balance analysis. BMC Systems Biology.
-

Basics of genome-scale metabolic modeling and applications on C1-utilization.
Kabimoldayev I, Nguyen ADuc, Yang L, Park S, Lee EYeol, Kim D. 2018. Basics of genome-scale metabolic modeling and applications on C1-utilization.. FEMS Microbiol Lett.
-

Genome-scale metabolic reconstructions of multiple Salmonella strains reveal serovar-specific metabolic traits.
Seif Y, Kavvas E, Lachance J-C, Yurkovich JT, Nuccio S-P, Fang X, Catoiu E, Raffatellu M, Palsson BO, Monk JM. 2018. Genome-scale metabolic reconstructions of multiple Salmonella strains reveal serovar-specific metabolic traits.. Nat Commun. 9(1):3771.
-

Evolution of gene knockout strains of E. coli reveal regulatory architectures governed by metabolism.
McCloskey D, Xu S, Sandberg TE, Brunk E, Hefner Y, Szubin R, Feist AM, Palsson BO. 2018. Evolution of gene knockout strains of E. coli reveal regulatory architectures governed by metabolism.. Nat Commun. 9(1):3796.
-

Gapless, Unambiguous Genome Sequence for Escherichia coli C, a Workhorse of Industrial Biology
Pekar JE, Phaneuf P, Szubin R, Palsson B, Feist A, Monk JM. Gapless, Unambiguous Genome Sequence for Escherichia coli C, a Workhorse of Industrial Biology. Microbiol Resour Announc. 2018 Oct 4;7(13). pii: e00890-18. doi:10.1128/MRA.00890-18. eCollection 2018 Oct. PubMed PMID: 30533692; PubMed Central PMCID: PMC6256561.
-

Metagenomics-Based, Strain-Level Analysis of Escherichia coli From a Time-Series of Microbiome Samples From a Crohn's Disease Patient
Fang X, Monk JM, Nurk S, Akseshina M, Zhu Q, Gemmell C, Gianetto-Hill C, Leung N, Szubin R, Sanders J, Beck PL, Li W, Sandborn WJ, Gray-Owen SD, Knight R, Allen-Vercoe E, Palsson BO, Smarr L. Metagenomics-Based, Strain-Level Analysis of Escherichia coli From a Time-Series of Microbiome Samples From a Crohn's Disease Patient. Front Microbiol. 2018 Oct 30;9:2559. doi: 10.3389/fmicb.2018.02559. eCollection 2018. PubMed PMID: 30425690; PubMed Central PMCID: PMC6218438.
-

Estimating Metabolic Equilibrium Constants: Progress and Future Challenges
Du B, Zielinski DC, Palsson BO. 2018. Estimating Metabolic Equilibrium Constants: Progress and Future Challenges. Trends in Biochemical Sciences.
-

Identification of growth-coupled production strains considering protein costs and kinetic variability
Dinh HV, King ZA, Palsson BO, Feist AM. 2018. Identification of growth-coupled production strains considering protein costs and kinetic variability. Metab Eng Commun. 2018 Oct 13;7:e00080. doi: 10.1016/j.mec.2018.e00080. eCollection 2018 Dec.
-

Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance
Kavvas ES, Catoiu E, Mih N, Yurkovich JT, Seif Y, Dillon N, Heckmann D, Anand A, Yang L, Nizet V, Monk JM, Palsson BO. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat Commun. 2018 Oct 17;9(1):4306. doi:10.1038/s41467-018-06634-y. PubMed PMID: 30333483; PubMed Central PMCID: PMC6193043.
-

Characterizing posttranslational modifications in prokaryotic metabolism using a multiscale workflow
Brunk E, Chang RL, Xia J, Hefzi H, Yurkovich JT, Kim D, Buckmiller E, Wang HH, Cho B-K, Yang C et al.. 2018. Characterizing posttranslational modifications in prokaryotic metabolism using a multiscale workflow. Proceedings of the National Academy of Sciences. 2018 Oct 23;115(43):11096-11101. doi: 10.1073/pnas.1811971115. Epub 2018 Oct 9.
-

Thermodynamic favorability and pathway yield as evolutionary tradeoffs in biosynthetic pathway choice
Du B, Zielinski DC, Monk JM, Palsson BO. 2018. Thermodynamic favorability and pathway yield as evolutionary tradeoffs in biosynthetic pathway choice. Proceedings of the National Academy of Sciences.
-

ALEdb 1.0: a database of mutations from adaptive laboratory evolution experimentation.
Phaneuf PV, Gosting D, Palsson BO, Feist AM. 2018. ALEdb 1.0: a database of mutations from adaptive laboratory evolution experimentation.. Nucleic Acids Res.
-

Predicting the metabolic capabilities of Synechococcus elongatus PCC 7942 adapted to different light regimes.
Broddrick JT, Welkie DG, Jallet D, Golden SS, Peers G, Palsson BO. 2018. Predicting the metabolic capabilities of Synechococcus elongatus PCC 7942 adapted to different light regimes.. Metab Eng.
-

Sugar-stimulated CO2 sequestration by the green microalga Chlorella vulgaris
Fu W, Gudmundsson S, Wichuk K, Palsson S, Palsson BO, Salehi-Ashtiani K, ólfsson ður. 2018. Sugar-stimulated CO2 sequestration by the green microalga Chlorella vulgaris. Science of The Total Environment. 654:275-283.
-

Revealing Key Determinants of Clonal Variation in Transgene Expression in Recombinant CHO Cells Using Targeted Genome Editing.
Lee JSeong, Park JHyoung, Ha TKwang, Samoudi M, Lewis NE, Palsson BO, Kildegaard HFaustrup, Lee G M. 2018. Revealing Key Determinants of Clonal Variation in Transgene Expression in Recombinant CHO Cells Using Targeted Genome Editing.. ACS Synth Biol.
-

Adaptation to the coupling of glycolysis to toxic methylglyoxal production in tpiA deletion strains of Escherichia coli requires synchronized and counterintuitive genetic changes.
McCloskey D, Xu S, Sandberg TE, Brunk E, Hefner Y, Szubin R, Feist AM, Palsson BO. 2018. Adaptation to the coupling of glycolysis to toxic methylglyoxal production in tpiA deletion strains of Escherichia coli requires synchronized and counterintuitive genetic changes.. Metab Eng.
-

The Staphylococcus aureus Two-Component System AgrAC Displays Four Distinct Genomic Arrangements That Delineate Genomic Virulence Factor Signatures
Choudhary KS, Mih N, Monk J, Kavvas E, Yurkovich JT, Sakoulas G, Palsson BO. 2018. The Staphylococcus aureus Two-Component System AgrAC Displays Four Distinct Genomic Arrangements That Delineate Genomic Virulence Factor Signatures. Frontiers in Microbiology. 2018 May 25;9:1082. doi: 10.3389/fmicb.2018.01082. eCollection 2018.
-

ChIP-exo interrogation of Crp, DNA, and RNAP holoenzyme interactions.
Latif H, Federowicz S, Ebrahim A, Tarasova J, Szubin R, Utrilla J, Zengler K, Palsson BO. 2018. ChIP-exo interrogation of Crp, DNA, and RNAP holoenzyme interactions.. PLoS One. 13(5):e0197272.
-

Laboratory evolution reveals regulatory and metabolic trade-offs of glycerol utilization in Saccharomyces cerevisiae.
Strucko T, Zirngibl K, Pereira F, Kafkia E, Mohamed ET, Rettel M, Stein F, Feist AM, Jouhten P, Patil KRaosaheb et al.. 2018. Laboratory evolution reveals regulatory and metabolic trade-offs of glycerol utilization in Saccharomyces cerevisiae.. Metab Eng. 47:73-82.
-

Systems biology as an emerging paradigm in transfusion medicine.
Yurkovich JT, Bordbar A, Sigurjónsson OE, Palsson BO. 2018. Systems biology as an emerging paradigm in transfusion medicine.. BMC Syst Biol. 12(1):31
-

Updated and standardized genome-scale reconstruction of Mycobacterium tuberculosis H37Rv, iEK1011, simulates flux states indicative of physiological conditions.
Kavvas ES, Seif Y, Yurkovich JT, Norsigian C, Poudel S, Greenwald WW, Ghatak S, Palsson BO, Monk JM. 2018. Updated and standardized genome-scale reconstruction of Mycobacterium tuberculosis H37Rv, iEK1011, simulates flux states indicative of physiological conditions.. BMC Syst Biol. 12(1):25.
-

Adaptive laboratory evolution resolves energy depletion to maintain high aromatic metabolite phenotypes in Escherichia coli strains lacking the Phosphotransferase System.
McCloskey D, Xu S, Sandberg TE, Brunk E, Hefner Y, Szubin R, Feist AM, Palsson BO. 2018. Adaptive laboratory evolution resolves energy depletion to maintain high aromatic metabolite phenotypes in Escherichia coli strains lacking the Phosphotransferase System.. Metab Eng.
-

Escherichia coli B2 strains prevalent in inflammatory bowel disease patients have distinct metabolic capabilities that enable colonization of intestinal mucosa.
Fang X, Monk JM, Mih N, Du B, Sastry AV, Kavvas E, Seif Y, Smarr L, Palsson BO. 2018. Escherichia coli B2 strains prevalent in inflammatory bowel disease patients have distinct metabolic capabilities that enable colonization of intestinal mucosa.. BMC Syst Biol. 12(1):66.
-

Temperature-Dependent Estimation of Gibbs Energies Using an Updated Group-Contribution Method.
Du B, Zhang Z, Grubner S, Yurkovich JT, Palsson BO, Zielinski DC. 2018. Temperature-Dependent Estimation of Gibbs Energies Using an Updated Group-Contribution Method.. Biophys J. 114(11):2691-2702.
-

COBRAme: A computational framework for genome-scale models of metabolism and gene expression.
Lloyd CJ, Ebrahim A, Yang L, King ZA, Catoiu E, O'Brien EJ, Liu JK, Palsson BO. 2018. COBRAme: A computational framework for genome-scale models of metabolism and gene expression.. PLoS Comput Biol. 14(7):e1006302.
-

Systems analysis of metabolism in platelet concentrates during storage in platelet additive solution.
Johannsson F, Guðmundsson S, Paglia G, Guðmundsson S, Palsson B, Sigurjónsson OE, Rolfsson O. 2018. Systems analysis of metabolism in platelet concentrates during storage in platelet additive solution.. Biochem J. 475(13):2225-2240.
-

Multiple optimal phenotypes overcome redox and glycolytic intermediate metabolite imbalances in knockout evolutions.
McCloskey D, Xu S, Sandberg TE, Brunk E, Hefner Y, Szubin R, Feist AM, Palsson BO. 2018. Multiple optimal phenotypes overcome redox and glycolytic intermediate metabolite imbalances in knockout evolutions.. Appl Environ Microbiol.
-

Systems assessment of transcriptional regulation on central carbon metabolism by Cra and CRP.
Kim D, Seo SWoo, Gao Y, Nam H, Guzman GI, Cho B-K, Palsson BO. 2018. Systems assessment of transcriptional regulation on central carbon metabolism by Cra and CRP.. Nucleic Acids Res.
-

Modeling the multi-scale mechanisms of macromolecular resource allocation
Yang L, Yurkovich JT, King ZA, Palsson BO. 2018. Modeling the multi-scale mechanisms of macromolecular resource allocation. Current Opinion in Microbiology. 45:8-15.
-

Functional interrogation of Plasmodium genus metabolism identifies species- and stage-specific differences in nutrient essentiality and drug targeting.
Abdel-Haleem AM, Hefzi H, Mineta K, Gao X, Gojobori T, Palsson BO, Lewis NE, Jamshidi N. 2018. Functional interrogation of Plasmodium genus metabolism identifies species- and stage-specific differences in nutrient essentiality and drug targeting.. PLoS Comput Biol. 14(1):e1005895.
-

ssbio: A Python Framework for Structural Systems Biology
Mih N, Brunk E, Chen K, Catoiu E, Sastry A, Kavvas E, Monk JM, Zhang Z, Palsson BO. 2018. ssbio: A Python Framework for Structural Systems Biology. Bioinformatics.
-

Genome-scale analysis of Methicillin-resistant Staphylococcus aureus USA300 reveals a tradeoff between pathogenesis and drug resistance.
Choe D, Szubin R, Dahesh S, Cho S, Nizet V, Palsson B, Cho B-K. 2018. Genome-scale analysis of Methicillin-resistant Staphylococcus aureus USA300 reveals a tradeoff between pathogenesis and drug resistance.. Sci Rep. 8(1):2215.
-

Quantitative -omic data empowers bottom-up systems biology
Yurkovich JT, Palsson BO. 2018. Quantitative -omic data empowers bottom-up systems biology. Current Opinion in Biotechnology. 51:130-136.
-

Recon3D enables a three-dimensional view of gene variation in human metabolism.
Brunk E, Sahoo S, Zielinski DC, Altunkaya A, Dräger A, Mih N, Gatto F, Nilsson A, Gonzalez GAndres Pre, Aurich MKathrin et al.. 2018. Recon3D enables a three-dimensional view of gene variation in human metabolism.. Nat Biotechnol.
-

Dataset on economic analysis of mass production of algae in LED-based photobioreactors.
Fu W, Gudmundsson S, Wichuk K, Palsson S, Palsson BO, Salehi-Ashtiani K, Brynjólfsson S. 2018. Dataset on economic analysis of mass production of algae in LED-based photobioreactors.. Data Brief. 22:137-139.
-

Reframing gene essentiality in terms of adaptive flexibility.
Guzman GI, Olson CA, Hefner Y, Phaneuf PV, Catoiu E, Crepaldi LB, Micas LGoldschmid, Palsson BO, Feist AM. 2018. Reframing gene essentiality in terms of adaptive flexibility.. BMC Syst Biol. 12(1):143.
-

Modeling genome-wide enzyme evolution predicts strong epistasis underlying catalytic turnover rates
Heckmann D, Zielinski DC, Palsson BO. 2018. Modeling genome-wide enzyme evolution predicts strong epistasis underlying catalytic turnover rates. Nature Communications. 9
-

Machine learning applied to enzyme turnover numbers reveals protein structural correlates and improves metabolic models
Heckmann D, Lloyd CJ, Mih N, Ha Y, Zielinski DC, Haiman ZB, Desouki AAmer, Lercher MJ, Palsson BO. 2018. Machine learning applied to enzyme turnover numbers reveals protein structural correlates and improves metabolic models. Nature Communications. 9
-

Network-level allosteric effects are elucidated by detailing how ligand-binding events modulate utilization of catalytic potentials.
Yurkovich JT, Alcantar MA, Haiman ZB, Palsson BO. 2018. Network-level allosteric effects are elucidated by detailing how ligand-binding events modulate utilization of catalytic potentials.. PLoS Comput Biol. 14(8):e1006356.
-

A unified resource for transcriptional regulation in Escherichia coli K-12 incorporating high-throughput-generated binding data into RegulonDB version 10.0
Santos-Zavaleta A, Sánchez-Pérez M, Salgado H, Vázquez-Ramírez DA, Gama-Castro S, Tierrafría VH, Busby SJW, Aquino P, Fang X, Palsson BO et al.. 2018. A unified resource for transcriptional regulation in Escherichia coli K-12 incorporating high-throughput-generated binding data into RegulonDB version 10.0. BMC Biology. 16
-

Growth Adaptation of gnd and sdhCB Escherichia coli Deletion Strains Diverges From a Similar Initial Perturbation of the Transcriptome
McCloskey D, Xu S, Sandberg TE, Brunk E, Hefner Y, Szubin R, Feist AM, Palsson BO. 2018. Growth Adaptation of gnd and sdhCB Escherichia coli Deletion Strains Diverges From a Similar Initial Perturbation of the Transcriptome. Frontiers in Microbiology.
-

Systematic discovery of uncharacterized transcription factors in Escherichia coli K-12 MG1655.
Gao Y, Yurkovich JT, Seo SWoo, Kabimoldayev I, Dräger A, Chen K, Sastry AV, Fang X, Mih N, Yang L et al.. 2018. Systematic discovery of uncharacterized transcription factors in Escherichia coli K-12 MG1655.. Nucleic Acids Res.
-

High-Level dCas9 Expression Induces Abnormal Cell Morphology in Escherichia coli.
Cho S, Choe D, Lee E, Kim SChang, Palsson B, Cho B-K. 2018. High-Level dCas9 Expression Induces Abnormal Cell Morphology in Escherichia coli.. ACS Synth Biol. 7(4):1085-1094.
-

iCN718, an Updated and Improved Genome-Scale Metabolic Network Reconstruction of Acinetobacter baumannii AYE
Norsigian CJ, Kavvas E, Seif Y, Palsson BO, Monk JM. 2018. iCN718, an Updated and Improved Genome-Scale Metabolic Network Reconstruction of Acinetobacter baumannii AYE. Frontiers in Genetics. 9
-

Global transcriptional regulatory network for Escherichia coli robustly connects gene expression to transcription factor activities.
Fang X, Sastry A, Mih N, Kim D, Tan J, Yurkovich JT, Lloyd CJ, Gao Y, Yang L, Palsson BO. 2017. Global transcriptional regulatory network for Escherichia coli robustly connects gene expression to transcription factor activities.. Proc Natl Acad Sci U S A.
-

Fast growth phenotype of E. coli K-12 from adaptive laboratory evolution does not require intracellular flux rewiring.
Long CP, Gonzalez JE, Feist AM, Palsson BO, Antoniewicz MR. 2017. Fast growth phenotype of E. coli K-12 from adaptive laboratory evolution does not require intracellular flux rewiring.. Metab Eng.
-

Quantitative time-course metabolomics in human red blood cells reveal the temperature dependence of human metabolic networks.
Yurkovich JT, Zielinski DC, Yang L, Paglia G, Rolfsson O, Sigurjónsson OE, Broddrick JT, Bordbar A, Wichuk K, Brynjólfsson S et al.. 2017. Quantitative time-course metabolomics in human red blood cells reveal the temperature dependence of human metabolic networks.. J Biol Chem.
-

iML1515, a knowledgebase that computes Escherichia coli traits.
Monk JM, Lloyd CJ, Brunk E, Mih N, Sastry A, King Z, Takeuchi R, Nomura W, Zhang Z, Mori H et al.. 2017. iML1515, a knowledgebase that computes Escherichia coli traits.. Nat Biotechnol. 35(10):904-908.
-

Thermosensitivity of growth is determined by chaperone-mediated proteome reallocation
Chen K, Gao Y, Mih N, Brien EJ ’, Yang L, Palsson BO. 2017. Thermosensitivity of growth is determined by chaperone-mediated proteome reallocation. Proceedings of the National Academy of Sciences. 377121391183:201705524.
-

A Padawan Programmer’s Guide to Developing Software Libraries
Yurkovich JT, Yurkovich BJ, Draeger A, Palsson BO, King ZA. 2017. A Padawan Programmer’s Guide to Developing Software Libraries. Cell Systems.
-

Antibiotic-Induced Changes to the Host Metabolic Environment Inhibit Drug Efficacy and Alter Immune Function.
Yang JH, Bhargava P, McCloskey D, Mao N, Palsson BO, Collins JJ. 2017. Antibiotic-Induced Changes to the Host Metabolic Environment Inhibit Drug Efficacy and Alter Immune Function.. Cell Host Microbe.
-

Generation of a platform strain for ionic liquid tolerance using adaptive laboratory evolution.
Mohamed ET, Wang S, Lennen RM, Herrgard MJ, Simmons BA, Singer SW, Feist AM. 2017. Generation of a platform strain for ionic liquid tolerance using adaptive laboratory evolution.. Microb Cell Fact. 16(1):204.
-

Expanding The Computable Reactome In Pseudomonas putida Reveals Metabolic Cycles Providing Robustness
Nogales J, Gudmundsson S, Duque E., Ramos JLewis, Palsson BO. 2017. Expanding The Computable Reactome In Pseudomonas putida Reveals Metabolic Cycles Providing Robustness. bioRxiv.
-

Revealing genome-scale transcriptional regulatory landscape of OmpR highlights its expanded regulatory roles under osmotic stress in Escherichia coli K-12 MG1655.
Seo SWoo, Gao Y, Kim D, Szubin R, Yang J, Cho B-K, Palsson BO. 2017. Revealing genome-scale transcriptional regulatory landscape of OmpR highlights its expanded regulatory roles under osmotic stress in Escherichia coli K-12 MG1655.. Sci Rep. 7(1):2181.
-

Metabolomics comparison of red cells stored in four additive solutions reveals differences in citrate anticoagulant permeability and metabolism.
Rolfsson Ó, Sigurjonsson ÓE, Magnusdottir M, Johannsson F, Paglia G, Guðmundsson S, Bordbar A, Palsson S, Brynjólfsson S, Guðmundsson S et al.. 2017. Metabolomics comparison of red cells stored in four additive solutions reveals differences in citrate anticoagulant permeability and metabolism.. Vox Sang.
-

Biomarkers are used to predict quantitative metabolite concentration profiles in human red blood cells.
Yurkovich JT, Yang L, Palsson BO. 2017. Biomarkers are used to predict quantitative metabolite concentration profiles in human red blood cells.. PLoS Comput Biol. 13(3):e1005424.
-

Systems biology analysis of drivers underlying hallmarks of cancer cell metabolism.
Zielinski DC, Jamshidi N, Corbett AJ, Bordbar A, Thomas A, Palsson BO. 2017. Systems biology analysis of drivers underlying hallmarks of cancer cell metabolism.. Sci Rep. 7:41241.
-

Whole-Genome Sequencing of Invasion-Resistant Cells Identifies Laminin α2 as a Host Factor for Bacterial Invasion.
van Wijk XM, Döhrmann S, Hallström BM, Li S, Voldborg BG, Meng BX, McKee KK, van Kuppevelt TH, Yurchenco PD, Palsson BO et al.. 2017. Whole-Genome Sequencing of Invasion-Resistant Cells Identifies Laminin α2 as a Host Factor for Bacterial Invasion.. MBio. 8(1)
-

Integrated Regulatory and Metabolic Networks of the Marine Diatom Phaeodactylum tricornutum Predict the Response to Rising CO2 Levels.
Levering J, Dupont CL, Allen AE, Palsson BO, Zengler K. 2017. Integrated Regulatory and Metabolic Networks of the Marine Diatom Phaeodactylum tricornutum Predict the Response to Rising CO2 Levels.. mSystems. 2(1)
-

COBRAme: A Computational Framework for Building and Manipulating Models of Metabolism and Gene Expression
Lloyd CJ, Ebrahim A, Yang L, King ZA, Catoiu E, O'Brien EJ, Liu JK, Palsson BO. 2017. COBRAme: A Computational Framework for Building and Manipulating Models of Metabolism and Gene Expression. bioRxiv.
-

Topological and Kinetic Determinants of the Modal Matrices of Dynamic Models of Metabolism
Du B, Zielinski DC, Palsson BO. 2017. Topological and Kinetic Determinants of the Modal Matrices of Dynamic Models of Metabolism. bioRxiv.
-

A Model for Designing Adaptive Laboratory Evolution Experiments.
LaCroix RA, Palsson BO, Feist AM. 2017. A Model for Designing Adaptive Laboratory Evolution Experiments.. Appl Environ Microbiol.
-

Metabolic Models of Protein Allocation Call for the Kinetome.
Nilsson A, Nielsen J, Palsson BO. 2017. Metabolic Models of Protein Allocation Call for the Kinetome.. Cell Syst. 5(6):538-541.
-

Dissecting the genetic and metabolic mechanisms of adaptation to the knockout of a major metabolic enzyme in Escherichia coli.
Long CP, Gonzalez JE, Feist AM, Palsson BO, Antoniewicz MR. 2017. Dissecting the genetic and metabolic mechanisms of adaptation to the knockout of a major metabolic enzyme in Escherichia coli.. Proc Natl Acad Sci U S A.
-

Mannose and fructose metabolism in red blood cells during cold storage in SAGM.
Rolfsson O, Johannsson F, Magnúsdóttir M, Paglia G, Sigurjónsson OE, Bordbar A, Palsson S, Brynjólfsson S, Guðmundsson S, Palsson B. 2017. Mannose and fructose metabolism in red blood cells during cold storage in SAGM.. Transfusion.
-

Underground metabolism: network-level perspective and biotechnological potential
Notebaart RA, Kintses álint, Feist AM, Papp ázs. 2017. Underground metabolism: network-level perspective and biotechnological potential. Current Opinion in Biotechnology. 49:108-114.
-

Utilizing biomarkers to forecast quantitative metabolite concentration profiles in human red blood cells
Yurkovich JT, Yang L, Palsson BO. 2017. Utilizing biomarkers to forecast quantitative metabolite concentration profiles in human red blood cells. 2017 IEEE Conference on Control Technology and Applications (CCTA).
-

Laboratory Evolution to Alternating Substrate Environments Yields Distinct Phenotypic and Genetic Adaptive Strategies.
Sandberg TE, Lloyd CJ, Palsson BO, Feist AM. 2017. Laboratory Evolution to Alternating Substrate Environments Yields Distinct Phenotypic and Genetic Adaptive Strategies.. Appl Environ Microbiol.
-

Machine Learning in Computational Biology to Accelerate High-Throughput Protein Expression.
Sastry A, Monk J, Tegel H, Uhlen M, Palsson BO, Rockberg J, Brunk E. 2017. Machine Learning in Computational Biology to Accelerate High-Throughput Protein Expression.. Bioinformatics.
-

Elucidating dynamic metabolic physiology through network integration of quantitative time-course metabolomics.
Bordbar A, Yurkovich JT, Paglia G, Rolfsson O, Sigurjónsson OE, Palsson BO. 2017. Elucidating dynamic metabolic physiology through network integration of quantitative time-course metabolomics.. Sci Rep. 7:46249.
-

A Phaeodactylum tricornutum literature database for interactive annotation of content
Gallina AA, Layer M, King ZA, Levering J, Palsson BO, Zengler K, Peers G. 2016. A Phaeodactylum tricornutum literature database for interactive annotation of content. Algal Research. 18:241-243.
-

Multi-omics Quantification of Species Variation of Escherichia coli Links Molecular Features with Strain Phenotypes.
Monk JM, Koza A, Campodonico MA, Machado D, Seoane JMiguel, Palsson BO, Herrgard MJ, Feist AM. 2016. Multi-omics Quantification of Species Variation of Escherichia coli Links Molecular Features with Strain Phenotypes.. Cell Syst.
-

solveME: fast and reliable solution of nonlinear ME models.
Yang L, Ma D, Ebrahim A, Lloyd CJ, Saunders MA, Palsson BO. 2016. solveME: fast and reliable solution of nonlinear ME models.. BMC Bioinformatics. 17(1):391.
-

Systems assessment of transcriptional regulation on central carbon metabolism by Cra and CRP
Kim D, Seo SWoo, Nam H, Guzman GI, Gao Y, Palsson BO. 2016. Systems assessment of transcriptional regulation on central carbon metabolism by Cra and CRP. bioRxiv.
-

Multi-omic data integration enables discovery of hidden biological regularities.
Ebrahim A, Brunk E, Tan J, O'Brien EJ, Kim D, Szubin R, Lerman JA, Lechner A, Sastry A, Bordbar A et al.. 2016. Multi-omic data integration enables discovery of hidden biological regularities.. Nat Commun. 7:13091.
-

Increased production of L-serine in Escherichia coli through Adaptive Laboratory Evolution.
Mundhada H, Seoane JM, Schneider K, Koza A, Christensen HB, Klein T, Phaneuf PV, Herrgard M, Feist AM, Nielsen AT. 2016. Increased production of L-serine in Escherichia coli through Adaptive Laboratory Evolution.. Metab Eng. 39:141-150.
-

Reliable and efficient solution of genome-scale models of Metabolism and macromolecular Expression.
Ma D, Yang L, Fleming RMT, Thiele I, Palsson BO, Saunders MA. 2016. Reliable and efficient solution of genome-scale models of Metabolism and macromolecular Expression.. Sci Rep. 7:40863.
-

A Consensus Genome-scale Reconstruction of Chinese Hamster Ovary Cell Metabolism.
Hefzi H, Ang KSiong, Hanscho M, Bordbar A, Ruckerbauer D, Lakshmanan M, Orellana CA, Baycin-Hizal D, Huang Y, Ley D et al.. 2016. A Consensus Genome-scale Reconstruction of Chinese Hamster Ovary Cell Metabolism.. Cell Syst. 3(5):434-443.e8.
-

The aldehyde dehydrogenase, AldA, is essential for L-1,2-propanediol utilization in laboratory-evolved Escherichia coli
Aziz R.K., Monk J.M., Andrews K.A., Nhan J., Khaw V.J., Wong H., Palsson B.O., Charusanti P.. 2016. The aldehyde dehydrogenase, AldA, is essential for L-1,2-propanediol utilization in laboratory-evolved Escherichia coli. Microbiological Research. 194:47-52.
-

Citrate metabolism in red blood cells stored in additive solution-3.
D'Alessandro A, Nemkov T, Yoshida T, Bordbar A, Palsson BO, Hansen KC. 2016. Citrate metabolism in red blood cells stored in additive solution-3.. Transfusion. 57(2):325-336.
-

Genome-Scale Model Reveals Metabolic Basis of Biomass Partitioning in a Model Diatom.
Levering J, Broddrick J, Dupont CL, Peers G, Beeri K, Mayers J, Gallina AA, Allen AE, Palsson BO, Zengler K. 2016. Genome-Scale Model Reveals Metabolic Basis of Biomass Partitioning in a Model Diatom.. PLoS One. 11(5):e0155038.
-

What do cells actually want?
Feist AM, Palsson BO. 2016. What do cells actually want? Genome Biol. 17(1):110.
-

Characterizing Strain Variation in Engineered E. coli Using a Multi-Omics-Based Workflow.
Brunk E, George KW, Alonso-Gutierrez J, Thompson M, Baidoo E, Wang G, Petzold CJ, McCloskey D, Monk J, Yang L et al.. 2016. Characterizing Strain Variation in Engineered E. coli Using a Multi-Omics-Based Workflow.. Cell Syst.
-

Quantitative feature extraction from the Chinese hamster ovary bioprocess bibliome using a novel meta-analysis workflow.
Golabgir A, Gutierrez JM, Hefzi H, Li S, Palsson BO, Herwig C, Lewis NE. 2016. Quantitative feature extraction from the Chinese hamster ovary bioprocess bibliome using a novel meta-analysis workflow.. Biotechnol Adv.
-

A modeling method for increased precision and scope of directly measurable fluxes at a genome-scale.
McCloskey D, Young JD, Xu S, Palsson BØ, Feist AM. 2016. A modeling method for increased precision and scope of directly measurable fluxes at a genome-scale.. Anal Chem.
-

Evolution of E. coli on [U-13C]Glucose Reveals a Negligible Isotopic Influence on Metabolism and Physiology.
Sandberg TE, Long CP, Gonzalez JE, Feist AM, Antoniewicz MR, Palsson BO. 2016. Evolution of E. coli on [U-13C]Glucose Reveals a Negligible Isotopic Influence on Metabolism and Physiology.. PLoS One. 11(3):e0151130.
-

Systems biology of the structural proteome.
Brunk E, Mih N, Monk J, Zhang Z, O'Brien EJ, Bliven SE, Chen K, Chang RL, Bourne PE, Palsson BO. 2016. Systems biology of the structural proteome.. BMC Syst Biol. 10(1):26.
-

Quantification and Classification of E. coli Proteome Utilization and Unused Protein Costs across Environments.
O'Brien EJ, Utrilla J, Palsson BO. 2016. Quantification and Classification of E. coli Proteome Utilization and Unused Protein Costs across Environments.. PLoS Comput Biol. 12(6):e1004998.
-

Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity.
Bosi E, Monk JM, Aziz RK, Fondi M, Nizet V, Palsson BØ. 2016. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity.. Proc Natl Acad Sci U S A.
-

Evaluation of rate law approximations in bottom-up kinetic models of metabolism.
Du B, Zielinski DC, Kavvas ES, Dräger A, Tan J, Zhang Z, Ruggiero KE, Arzumanyan GA, Palsson BO. 2016. Evaluation of rate law approximations in bottom-up kinetic models of metabolism.. BMC Syst Biol. 10(1):40.
-

A Multi-scale Computational Platform to Mechanistically Assess the Effect of Genetic Variation on Drug Responses in Human Erythrocyte Metabolism.
Mih N, Brunk E, Bordbar A, Palsson BO. 2016. A Multi-scale Computational Platform to Mechanistically Assess the Effect of Genetic Variation on Drug Responses in Human Erythrocyte Metabolism.. PLoS Comput Biol. 12(7):e1005039.
-

Construction and Evaluation of an Organic Anion Transporter 1 (OAT1)-Centered Metabolic Network.
Liu HC, Jamshidi N, Chen Y, Eraly SA, Cho SYee, Bhatnagar V, Wu W, Bush KT, Abagyan R, Palsson BO et al.. 2016. Construction and Evaluation of an Organic Anion Transporter 1 (OAT1)-Centered Metabolic Network.. J Biol Chem.
-

Identified metabolic signature for assessing red blood cell unit quality is associated with endothelial damage markers and clinical outcomes.
Bordbar A, Johansson PI, Paglia G, Harrison SJ, Wichuk K, Magnúsdóttir M, Valgeirsdottir S, Gybel-Brask M, Ostrowski SR, Palsson S et al.. 2016. Identified metabolic signature for assessing red blood cell unit quality is associated with endothelial damage markers and clinical outcomes.. Transfusion.
-

BiGG Models: A platform for integrating, standardizing and sharing genome-scale models.
King ZA, Lu J, Dräger A, Miller P, Federowicz S, Lerman JA, Ebrahim A, Palsson BO, Lewis NE. 2016. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models.. Nucleic Acids Res.
-
Solving puzzles with missing pieces: The power of systems biology
Yurkovich J.T., Palsson B.O.. 2016. Solving puzzles with missing pieces: The power of systems biology. Proceedings of the IEEE. 104(1)
-

What Makes a Bacterial Species Pathogenic?:Comparative Genomic Analysis of the Genus Leptospira.
Fouts DE, Matthias MA, Adhikarla H, Adler B, Amorim-Santos L, Berg DE, Bulach D, Buschiazzo A, Chang Y-F, Galloway RL et al.. 2016. What Makes a Bacterial Species Pathogenic?:Comparative Genomic Analysis of the Genus Leptospira. PLoS Negl Trop Dis. 10(2):e0004403.
-

Unique attributes of cyanobacterial metabolism revealed by improved genome-scale metabolic modeling and essential gene analysis.
Broddrick JT, Rubin BE, Welkie DG, Du N, Mih N, Diamond S, Lee JJ, Golden SS, Palsson BO. 2016. Unique attributes of cyanobacterial metabolism revealed by improved genome-scale metabolic modeling and essential gene analysis.. Proc Natl Acad Sci U S A. 113(51):E8344-E8353.
-

Literature mining supports a next-generation modeling approach to predict cellular byproduct secretion.
King ZA, O'Brien EJ, Feist AM, Palsson BO. 2016. Literature mining supports a next-generation modeling approach to predict cellular byproduct secretion.. Metab Eng.
-

ChIP-exo interrogation of Crp, DNA, and RNAP holoenzyme interactions
Latif H, Federowicz S, Ebrahim A, Tarasova J, Szubin R, Utrilla J, Zengler K, Palsson BO. 2016. ChIP-exo interrogation of Crp, DNA, and RNAP holoenzyme interactions. bioRxiv.
-

Biomarkers defining the metabolic age of red blood cells during cold storage.
Paglia G, D'Alessandro A, Rolfsson O, Sigurjónsson OE, Bordbar A, Palsson S, Nemkov T, Hansen KC, Gudmundsson S, Palsson BO. 2016. Biomarkers defining the metabolic age of red blood cells during cold storage.. Blood.
-

Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking.
Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen D D, Watrous J, Kapono CA, Luzzatto-Knaan T et al.. 2016. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking.. Nat Biotechnol. 34(8):828-837.
-

Metabolic fate of adenine in red blood cells during storage in SAGM solution.
Paglia G, Sigurjónsson OE, Bordbar A, Rolfsson O, Magnúsdóttir M, Palsson S, Wichuk K, Gudmundsson S, Palsson BO. 2016. Metabolic fate of adenine in red blood cells during storage in SAGM solution.. Transfusion.
-

Global Rebalancing of Cellular Resources by Pleiotropic Point Mutations Illustrates a Multi-scale Mechanism of Adaptive Evolution.
Utrilla J, O'Brien EJ, Chen K, McCloskey D, Cheung J, Wang H, Armenta-Medina D, Feist AM, Palsson BO. 2016. Global Rebalancing of Cellular Resources by Pleiotropic Point Mutations Illustrates a Multi-scale Mechanism of Adaptive Evolution.. Cell Syst. 2(4):260-71.
-

Acidithiobacillus ferrooxidans's comprehensive model driven analysis of the electron transfer metabolism and synthetic strain design for biomining applications
Campodonico MA, Vaisman D, Castro JF, Razmilic V, Mercado F, Andrews BA, Feist AM, Asenjo JA. 2016. Acidithiobacillus ferrooxidans's comprehensive model driven analysis of the electron transfer metabolism and synthetic strain design for biomining applications. Metabolic Engineering Communications. 3
-

Model-driven discovery of synergistic inhibitors against E. coli and S. enterica serovar Typhimurium targeting a novel synthetic lethal pair, aldA and prpC.
Aziz RK, Khaw VL, Monk JM, Brunk E, Lewis R, Loh SI, Mishra A, Nagle AA, Satyanarayana C, Dhakshinamoorthy S et al.. 2015. Model-driven discovery of synergistic inhibitors against E. coli and S. enterica serovar Typhimurium targeting a novel synthetic lethal pair, aldA and prpC.. Front Microbiol. 6:958.
-
Do genome-scale models need exact solvers or clearer standards?
Ebrahim A, Almaas E, Bauer E, Bordbar A, Burgard AP, Chang RL, Dräger A, Famili I, Feist AM, Fleming RMt et al.. 2015. Do genome-scale models need exact solvers or clearer standards? Mol Syst Biol. 11(10):831.
-

Personalized Whole-Cell Kinetic Models of Metabolism for Discovery in Genomics and Pharmacodynamics
Bordbar A., McCloskey D., Zielinski D.C., Sonnenschein N., Jamshidi N., Palsson B.O.. 2015. Personalized Whole-Cell Kinetic Models of Metabolism for Discovery in Genomics and Pharmacodynamics. Cell Systems. 1:283-292.
-

SBMLsqueezer 2: context-sensitive creation of kinetic equations in biochemical networks.
Dräger A, Zielinski DC, Keller R, Rall M, Eichner J, Palsson BO, Zell A. 2015. SBMLsqueezer 2: context-sensitive creation of kinetic equations in biochemical networks.. BMC Syst Biol. 9(1):68.
-

A Markov chain model for N-linked protein glycosylation - towards a low-parameter tool for model-driven glycoengineering.
Spahn PN, Hansen AH, Hansen HG, Arnsdorf J, Kildegaard HF, Lewis NE. 2015. A Markov chain model for N-linked protein glycosylation - towards a low-parameter tool for model-driven glycoengineering.. Metab Eng.
-

Systems biology-guided identification of synthetic lethal gene pairs and its potential use to discover antibiotic combinations
Aziz R.K., Monk J.M., Lewis R.M., Loh S.I., Mishra A., Nagle A.A., Satyanarayana C., Dhakshinamoorthy S., Luche M., Kitchen D.B. et al.. 2015. Systems biology-guided identification of synthetic lethal gene pairs and its potential use to discover antibiotic combinations. Scientific Reports. doi:10.1038/srep16025
-

Networks of energetic and metabolic interactions define dynamics in microbial communities.
Embree M, Liu JK, Al-Bassam MM, Zengler K. 2015. Networks of energetic and metabolic interactions define dynamics in microbial communities.. Proc Natl Acad Sci U S A.
-

A logical data representation framework for electricity-driven bioproduction processes.
Patil SA, Gildemyn S, Pant D, Zengler K, Logan BE, Rabaey K. 2015. A logical data representation framework for electricity-driven bioproduction processes.. Biotechnol Adv. 33(6 Pt 1):736-44.
-

Using Genome-scale Models to Predict Biological Capabilities.
O'Brien EJ, Monk JM, Palsson BO. 2015. Using Genome-scale Models to Predict Biological Capabilities.. Cell. 161(5):971-987.
-

Unraveling interactions in microbial communities - from co-cultures to microbiomes.
Tan J, Zuniga C, Zengler K. 2015. Unraveling interactions in microbial communities - from co-cultures to microbiomes.. J Microbiol. 53(5):295-305.
-

Decoding the jargon of bottom-up metabolic systems biology.
Rolfsson O, Palsson BO. 2015. Decoding the jargon of bottom-up metabolic systems biology.. Bioessays.
-

The architecture of ArgR-DNA complexes at the genome-scale in Escherichia coli.
Cho S, Cho Y-B, Kang TJin, Kim SChang, Palsson B, Cho B-K. 2015. The architecture of ArgR-DNA complexes at the genome-scale in Escherichia coli.. Nucleic Acids Res.
-

Multidimensional Analytical Approach Based on UHPLC-UV-Ion Mobility-MS for the Screening of Natural Pigments.
Pacini T, Fu W, Gudmundsson S, A Chiaravalle E, Brynjolfson S, Palsson BO, Astarita G, Paglia G. 2015. Multidimensional Analytical Approach Based on UHPLC-UV-Ion Mobility-MS for the Screening of Natural Pigments.. Anal Chem. 87(5):2593-9.
-

Prediction of intracellular metabolic states from extracellular metabolomic data.
Aurich MK, Paglia G, Rolfsson O, Hrafnsdóttir S, Magnúsdóttir M, Stefaniak MM, Palsson BØ, Fleming RMT, Thiele I. 2015. Prediction of intracellular metabolic states from extracellular metabolomic data.. Metabolomics. 11(3):603-619.
-

How to set up collaborations between academia and industrial biotech companies.
Pronk JT, Lee S Y, Lievense J, Pierce J, Palsson B, Uhlen M, Nielsen J. 2015. How to set up collaborations between academia and industrial biotech companies.. Nat Biotechnol. 33(3):237-40.
-

Next-generation genome-scale models for metabolic engineering.
King ZA, Lloyd CJ, Feist AM, Palsson BO. 2015. Next-generation genome-scale models for metabolic engineering.. Curr Opin Biotechnol. 35C:23-29.
-

CONDENSED VERSION--Pharmacogenomic and clinical data link non-pharmacokinetic metabolic dysregulation to drug side effect pathogenesis
Zielinski DC, Filipp FV, Bordbar A, Jensen K, Smith JW, Herrgard MJ, Mo ML, Palsson BØ. 2015. CONDENSED VERSION--Pharmacogenomic and clinical data link non-pharmacokinetic metabolic dysregulation to drug side effect pathogenesis.
-

A streamlined ribosome profiling protocol for the characterization of microorganisms.
Latif H, Szubin R, Tan J, Brunk E, Lechner A, Zengler K, Palsson BO. 2015. A streamlined ribosome profiling protocol for the characterization of microorganisms.. Biotechniques. 58(6):329-32.
-

JSBML 1.0: providing a smorgasbord of options to encode systems biology models.
Rodriguez N, Thomas A, Watanabe L, Vazirabad IY, Kofia V, Gómez HF, Mittag F, Matthes J, Rudolph J, Wrzodek F et al.. 2015. JSBML 1.0: providing a smorgasbord of options to encode systems biology models.. Bioinformatics.
-

Pharmacogenomic and clinical data link non-pharmacokinetic metabolic dysregulation to drug side effect pathogenesis.
Zielinski DC, Filipp FV, Bordbar A, Jensen K, Smith JW, Herrgard MJ, Mo ML, Palsson BO. 2015. Pharmacogenomic and clinical data link non-pharmacokinetic metabolic dysregulation to drug side effect pathogenesis.. Nat Commun. 6:7101.
-

Optimizing eukaryotic cell hosts for protein production through systems biotechnology and genome-scale modeling.
Gutierrez JM, Lewis NE. 2015. Optimizing eukaryotic cell hosts for protein production through systems biotechnology and genome-scale modeling.. Biotechnol J. 10(7):939-49.
-

Computing the functional proteome: recent progress and future prospects for genome-scale models.
O'Brien EJ, Palsson BO. 2015. Computing the functional proteome: recent progress and future prospects for genome-scale models.. Curr Opin Biotechnol. 34C:125-134.
-

Model-driven discovery of underground metabolic functions in Escherichia coli.
Guzman GI, Utrilla J, Nurk S, Brunk E, Monk JM, Ebrahim A, Palsson BO, Feist AM. 2015. Model-driven discovery of underground metabolic functions in Escherichia coli.. Proc Natl Acad Sci U S A.
-

A pH and solvent optimized reverse-phase ion-paring-LC–MS/MS method that leverages multiple scan-types for targeted absolute quantification of intracellular metabolites
McCloskey DM, Gangoiti JA, Palsson BO, Feist AM. 2015. A pH and solvent optimized reverse-phase ion-paring-LC–MS/MS method that leverages multiple scan-types for targeted absolute quantification of intracellular metabolites. Metabolomics. 10.1007/s11306-015-0790-y
-

MID Max: LC-MS/MS Method for Measuring the Precursor and Product Mass Isotopomer Distributions of Metabolic Intermediates and Cofactors for Metabolic Flux Analysis Applications.
McCloskey D, Young JD, Xu S, Palsson BO, Feist AM. 2015. MID Max: LC-MS/MS Method for Measuring the Precursor and Product Mass Isotopomer Distributions of Metabolic Intermediates and Cofactors for Metabolic Flux Analysis Applications.. Anal Chem.
-

Escher: A Web Application for Building, Sharing, and Embedding Data-Rich Visualizations of Biological Pathways.
King ZA, Dräger A, Ebrahim A, Sonnenschein N, Lewis NE, Palsson BO. 2015. Escher: A Web Application for Building, Sharing, and Embedding Data-Rich Visualizations of Biological Pathways.. PLoS Comput Biol. 11(8):e1004321.
-

Systems biology definition of the core proteome of metabolism and expression is consistent with high-throughput data.
Yang L, Tan J, O'Brien EJ, Monk JM, Kim D, Li HJ, Charusanti P, Ebrahim A, Lloyd CJ, Yurkovich JT et al.. 2015. Systems biology definition of the core proteome of metabolism and expression is consistent with high-throughput data.. Proc Natl Acad Sci U S A.
-

Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli.
Seo SWoo, Kim D, O'Brien EJ, Szubin R, Palsson BO. 2015. Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli.. Nat Commun. 6:7970.
-

Engineering of oleaginous organisms for lipid production.
Levering J, Broddrick J, Zengler K. 2015. Engineering of oleaginous organisms for lipid production.. Curr Opin Biotechnol. 36:32-39.
-

Adaptive Evolution of Thermotoga maritima Reveals Plasticity of the ABC Transporter Network.
Latif H, Sahin M, Tarasova J, Tarasova Y, Portnoy VA, Nogales J, Zengler K. 2015. Adaptive Evolution of Thermotoga maritima Reveals Plasticity of the ABC Transporter Network.. Appl Environ Microbiol. 81(16):5477-85.
-

Investigating Moorella thermoacetica metabolism with a genome-scale constraint-based metabolic model.
M Islam A, Zengler K, Edwards EA, Mahadevan R, Stephanopoulos G. 2015. Investigating Moorella thermoacetica metabolism with a genome-scale constraint-based metabolic model.. Integr Biol (Camb). 7(8):869-82.
-

Genome-wide Reconstruction of OxyR and SoxRS Transcriptional Regulatory Networks under Oxidative Stress in Escherichia coli K-12 MG1655.
Seo SWoo, Kim D, Szubin R, Palsson BO. 2015. Genome-wide Reconstruction of OxyR and SoxRS Transcriptional Regulatory Networks under Oxidative Stress in Escherichia coli K-12 MG1655.. Cell Rep.
-

A Systems Approach to Predict Oncometabolites via Context-Specific Genome-Scale Metabolic Networks.
Nam H, Campodonico M, Bordbar A, Hyduke DR, Kim S, Zielinski DC, Palsson BO. 2014. A Systems Approach to Predict Oncometabolites via Context-Specific Genome-Scale Metabolic Networks.. PLoS Comput Biol. 10(9):e1003837.
-

Reconstruction and modeling protein translocation and compartmentalization in Escherichia coli at the genome-scale.
Liu JK, Brien EJO, Lerman JA, Zengler K, Palsson BO, Feist AM. 2014. Reconstruction and modeling protein translocation and compartmentalization in Escherichia coli at the genome-scale.. BMC Syst Biol. 8(1):110.
-

Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli.
Seo SWoo, Kim D, Latif H, O'Brien EJ, Szubin R, Palsson BO. 2014. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli.. Nat Commun. 5:4910.
-

Systems glycobiology for glycoengineering.
Spahn PN, Lewis NE. 2014. Systems glycobiology for glycoengineering.. Curr Opin Biotechnol. 30C:218-224.
-
Supplementation of Saturated Long-chain Fatty Acids Maintains Intestinal Eubiosis and Reduces Ethanol-induced Liver Injury in Mice.
Chen P, Torralba M, Tan J, Embree M, Zengler K, Stärkel P, van Pijkeren J-P, DePew J, Loomba R, Ho SB et al.. 2014. Supplementation of Saturated Long-chain Fatty Acids Maintains Intestinal Eubiosis and Reduces Ethanol-induced Liver Injury in Mice.. Gastroenterology.
-
The Metabolic Impact of a NADH-producing Glucose-6-phosphate Dehydrogenase in Escherichia coli.
Olavarria K, De Ingeniis J, Zielinski DC, Fuentealba M, Muñoz R, McCloskey D, Feist AM, Cabrera R. 2014. The Metabolic Impact of a NADH-producing Glucose-6-phosphate Dehydrogenase in Escherichia coli.. Microbiology.
-

Discovery of key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal media using adaptive laboratory evolution.
LaCroix RA, Sandberg TE, O'Brien EJ, Utrilla J, Ebrahim A, Guzman GI, Szubin R, Palsson BO, Feist AM. 2014. Discovery of key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal media using adaptive laboratory evolution.. Appl Environ Microbiol. \
-

Comprehensive metabolomic study of platelets reveals the expression of discrete metabolic phenotypes during storage.
Paglia G, Sigurjónsson OE, Rolfsson O, Valgeirsdottir S, Hansen MBagge, Brynjólfsson S, Gudmundsson S, Palsson BO. 2014. Comprehensive metabolomic study of platelets reveals the expression of discrete metabolic phenotypes during storage.. Transfusion.
-

From random mutagenesis to systems biology in metabolic engineering of mammalian cells
Hefzi H., Lewis N.E.. 2014. From random mutagenesis to systems biology in metabolic engineering of mammalian cells. Pharmaceutical Bioprocessing. 2
-

Biochemical Characterization of Human Gluconokinase and the Proposed Metabolic Impact of Gluconic Acid as Determined by Constraint Based Metabolic Network Analysis.
Rohatgi N, Nielsen TKragh, Bjørn SPetersen, Axelsson I, Paglia G, Voldborg BGunnar, Palsson BO, Rolfsson O. 2014. Biochemical Characterization of Human Gluconokinase and the Proposed Metabolic Impact of Gluconic Acid as Determined by Constraint Based Metabolic Network Analysis.. PLoS One. 9(6):e98760.
-

Fast Swinnex filtration (FSF): a fast and robust sampling and extraction method suitable for metabolomics analysis of cultures grown in complex media
McCloskey DM, Utrilla J, Naviaux RK, Palsson BO, Feist AM. 2014. Fast Swinnex filtration (FSF): a fast and robust sampling and extraction method suitable for metabolomics analysis of cultures grown in complex media. Metabolomics. 10(10.1007/s11306-014-0686-2)
-

Generation of an atlas for commodity chemical production in Escherichia coli and a novel pathway prediction algorithm, GEM-Path.
Campodonico MA, Andrews BA, Asenjo JA, Palsson BO, Feist AM. 2014. Generation of an atlas for commodity chemical production in Escherichia coli and a novel pathway prediction algorithm, GEM-Path.. Metab Eng. 25C:140-158.
-

Evolution of Escherichia coli to 42 °C and Subsequent Genetic Engineering Reveals Adaptive Mechanisms and Novel Mutations.
Sandberg TE, Pedersen M, LaCroix RA, Ebrahim A, Bonde M, Herrgard MJ, Palsson BO, Sommer M, Feist AM. 2014. Evolution of Escherichia coli to 42 °C and Subsequent Genetic Engineering Reveals Adaptive Mechanisms and Novel Mutations.. Mol Biol Evol.
-

Multi-Tissue Computational Modeling Analyzes Pathophysiology of Type 2 Diabetes in MKR Mice.
Kumar A, Harrelson T, Lewis NE, Gallagher EJ, LeRoith D, Shiloach J, Betenbaugh MJ. 2014. Multi-Tissue Computational Modeling Analyzes Pathophysiology of Type 2 Diabetes in MKR Mice.. PLoS One. 9(7):e102319.
-

Minimal metabolic pathway structure is consistent with associated biomolecular interactions.
Bordbar A, Nagarajan H, Lewis NE, Latif H, Ebrahim A, Federowicz S, Schellenberger J, Palsson BO. 2014. Minimal metabolic pathway structure is consistent with associated biomolecular interactions.. Mol Syst Biol. 10(7):737.
-

Ion Mobility-Derived Collision Cross Section As an Additional Measure for Lipid Fingerprinting and Identification.
Paglia G, Angel P, Williams JP, Richardson K, Olivos HJ, J Thompson W, Menikarachchi L, Lai S, Walsh C, Moseley A et al.. 2014. Ion Mobility-Derived Collision Cross Section As an Additional Measure for Lipid Fingerprinting and Identification.. Anal Chem.
-

Improving collaboration by standardization efforts in systems biology.
Dräger A, Palsson BØ. 2014. Improving collaboration by standardization efforts in systems biology.. Front Bioeng Biotechnol. 2:61.
-

Metabolomic analysis of platelets during storage: a comparison between apheresis- and buffy coat-derived platelet concentrates.
Paglia G, Sigurjónsson OE, Rolfsson O, Hansen MBagge, Brynjólfsson S, Gudmundsson S, Palsson BO. 2014. Metabolomic analysis of platelets during storage: a comparison between apheresis- and buffy coat-derived platelet concentrates.. Transfusion.
-

A Gapless, Unambiguous Genome Sequence of the Enterohemorrhagic Escherichia coli O157:H7 Strain EDL933.
Latif H, Li HJ, Charusanti P, Palsson BØ, Aziz RK. 2014. A Gapless, Unambiguous Genome Sequence of the Enterohemorrhagic Escherichia coli O157:H7 Strain EDL933.. Genome Announc. 2(4)
-

Optimizing genome-scale network reconstructions.
Monk J, Nogales J, Palsson BO. 2014. Optimizing genome-scale network reconstructions.. Nat Biotechnol. 32(5):447-452.
-

Tracing Compartmentalized NADPH Metabolism in the Cytosol and Mitochondria of Mammalian Cells.
Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Heiden MGVander, Metallo CM. 2014. Tracing Compartmentalized NADPH Metabolism in the Cytosol and Mitochondria of Mammalian Cells.. Mol Cell.
-

Optimal cofactor swapping can increase the theoretical yield for chemical production in Escherichia coli and Saccharomyces cerevisiae.
King ZA, Feist AM. 2014. Optimal cofactor swapping can increase the theoretical yield for chemical production in Escherichia coli and Saccharomyces cerevisiae.. Metab Eng. 24C:117-128.
-

Predicting microbial growth.
Monk J, Palsson BO. 2014. Predicting microbial growth.. Science. 344(6191):1448-9.
-

Capsule deletion via a lamda-Red knockout system perturbs biofilm formation and fimbriae expression in Klebsiella pneumoniae MGH 78578.
Huang T-W, Lam I, Chang H-Y, Tsai S-F, Palsson BO, Charusanti P. 2014. Capsule deletion via a lamda-Red knockout system perturbs biofilm formation and fimbriae expression in Klebsiella pneumoniae MGH 78578.. BMC Res Notes. 7(1):13.
-

Global metabolic network reorganization by adaptive mutations allows fast growth of Escherichia coli on glycerol.
Cheng K-K, Lee B-S, Masuda T, Ito T, Ikeda K, Hirayama A, Deng L, Dong J, Shimizu K, Soga T et al.. 2014. Global metabolic network reorganization by adaptive mutations allows fast growth of Escherichia coli on glycerol.. Nat Commun. 5:3233.
-

Genome-scale reconstruction of the sigma factor network in Escherichia coli: topology and functional states.
Cho B-K, Kim D, Knight EM, Zengler K, Palsson BO. 2014. Genome-scale reconstruction of the sigma factor network in Escherichia coli: topology and functional states.. BMC Biol. 12(1):4.
-

Cofactory: Sequence-based prediction of cofactor specificity of Rossmann folds.
Geertz-Hansen HMarcus, Blom N, Feist AM, Brunak S, Petersen TNordahl. 2014. Cofactory: Sequence-based prediction of cofactor specificity of Rossmann folds.. Proteins.
-

Constraint-based models predict metabolic and associated cellular functions.
Bordbar A, Monk JM, King ZA, Palsson BO. 2014. Constraint-based models predict metabolic and associated cellular functions.. Nat Rev Genet. 15(2):107-20.
-

Engineering synergy in biotechnology.
Nielsen J, Fussenegger M, Keasling J, Lee S Y, Liao JC, Prather K, Palsson B. 2014. Engineering synergy in biotechnology.. Nat Chem Biol. 10(5):319-22.
-

Ion mobility derived collision cross sections to support metabolomics applications.
Paglia G, Williams JP, Menikarachchi L, J Thompson W, Tyldesley-Worster R, Halldórsson S, Rolfsson O, Moseley A, Grant D, Langridge J et al.. 2014. Ion mobility derived collision cross sections to support metabolomics applications.. Anal Chem. 86(8):3985-93.
-

Constraint-Based Modeling of Carbon Fixation and the Energetics of Electron Transfer in Geobacter metallireducens.
Feist AM, Nagarajan H, Rotaru A-E, Tremblay P-L, Zhang T, Nevin KP, Lovley DR, Zengler K. 2014. Constraint-Based Modeling of Carbon Fixation and the Energetics of Electron Transfer in Geobacter metallireducens.. PLoS Comput Biol. 10(4):e1003575.
-

Determining the control circuitry of redox metabolism at the genome-scale.
Federowicz S, Kim D, Ebrahim A, Lerman J, Nagarajan H, Cho B-K, Zengler K, Palsson B. 2014. Determining the control circuitry of redox metabolism at the genome-scale.. PLoS Genet. 10(4):e1004264.
-

Effects of abiotic stressors on lutein production in the green microalga Dunaliella salina.
Fu W, Paglia G, Magnúsdóttir M, Steinarsdóttir EA, Gudmundsson S, Palsson BO, Andrésson OS, Brynjólfsson S. 2014. Effects of abiotic stressors on lutein production in the green microalga Dunaliella salina.. Microb Cell Fact. 13(1):3.
-

Network reconstruction of platelet metabolism identifies metabolic signature for aspirin resistance.
Thomas A, Rahmanian S, Bordbar A, Palsson BØ, Jamshidi N. 2014. Network reconstruction of platelet metabolism identifies metabolic signature for aspirin resistance.. Sci Rep. 4:3925.
-

Single-cell genome and metatranscriptome sequencing reveal metabolic interactions of an alkane-degrading methanogenic community.
Embree M, Nagarajan H, Movahedi N, Chitsaz H, Zengler K. 2013. Single-cell genome and metatranscriptome sequencing reveal metabolic interactions of an alkane-degrading methanogenic community.. ISME J.
-

Antibacterial mechanisms identified through structural systems pharmacology.
Chang RL, Xie L, Bourne PE, Palsson BO. 2013. Antibacterial mechanisms identified through structural systems pharmacology.. BMC Syst Biol. 7(1):102.
-

Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction.
O'Brien EJ, Lerman JA, Chang RL, Hyduke DR, Palsson BØ. 2013. Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction.. Mol Syst Biol. 9:693.
-

A model-driven quantitative metabolomics analysis of aerobic and anaerobic metabolism in E. coli K-12 MG1655 that is biochemically and thermodynamically consistent.
McCloskey D, Gangoiti JA, King ZA, Naviaux RK, Barshop BA, Palsson BO, Feist AM. 2013. A model-driven quantitative metabolomics analysis of aerobic and anaerobic metabolism in E. coli K-12 MG1655 that is biochemically and thermodynamically consistent.. Biotechnol Bioeng.
-

Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites.
Hwang K-S, Kim H U, Charusanti P, Palsson BØ, Lee S Y. 2013. Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites.. Biotechnol Adv. 32(2):255-268.
-

Path2Models: large-scale generation of computational models from biochemical pathway maps.
Büchel F, Rodriguez N, Swainston N, Wrzodek C, Czauderna T, Keller R, Mittag F, Schubert M, Glont M, Golebiewski M et al.. 2013. Path2Models: large-scale generation of computational models from biochemical pathway maps.. BMC Syst Biol. 7:116.
-

Parkinson's disease: dopaminergic nerve cell model is consistent with experimental finding of increased extracellular transport of α-synuclein.
Büchel F, Saliger S, Dräger A, Hoffmann S, Wrzodek C, Zell A, Kahle PJ. 2013. Parkinson's disease: dopaminergic nerve cell model is consistent with experimental finding of increased extracellular transport of α-synuclein.. BMC Neurosci. 14:136.
-

Characterization and modelling of interspecies electron transfer mechanisms and microbial community dynamics of a syntrophic association.
Nagarajan H, Embree M, Rotaru A-E, Shrestha PM, Feist AM, Palsson BØ, Lovley DR, Zengler K. 2013. Characterization and modelling of interspecies electron transfer mechanisms and microbial community dynamics of a syntrophic association.. Nat Commun. 4:2809.
-

Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments.
Monk JM, Charusanti P, Aziz RK, Lerman JA, Premyodhin N, Orth JD, Feist AM, Palsson BØ. 2013. Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments.. Proc Natl Acad Sci U S A.
-

The systems biology simulation core algorithm.
Keller R, Dörr A, Tabira A, Funahashi A, Ziller MJ, Adams R, Rodriguez N, Le Novère N, Hiroi N, Planatscher H et al.. 2013. The systems biology simulation core algorithm.. BMC Syst Biol. 7:55.
-

TFpredict and SABINE: sequence-based prediction of structural and functional characteristics of transcription factors.
Eichner J, Topf F, Dräger A, Wrzodek C, Wanke D, Zell A. 2013. TFpredict and SABINE: sequence-based prediction of structural and functional characteristics of transcription factors.. PLoS One. 8(12):e82238.
-

SBML qualitative models: a model representation format and infrastructure to foster interactions between qualitative modelling formalisms and tools.
Chaouiya C, Bérenguier D, Keating SM, Naldi A, van Iersel MP, Rodriguez N, Dräger A, Büchel F, Cokelaer T, Kowal B et al.. 2013. SBML qualitative models: a model representation format and infrastructure to foster interactions between qualitative modelling formalisms and tools.. BMC Syst Biol. 7:135.
-

Transcriptional regulation of the carbohydrate utilization network in Thermotoga maritima.
Rodionov DA, Rodionova IA, Li X, Ravcheev DA, Tarasova Y, Portnoy VA, Zengler K, Osterman AL. 2013. Transcriptional regulation of the carbohydrate utilization network in Thermotoga maritima.. Front Microbiol. 4:244.
-

Optimizing Cofactor Specificity of Oxidoreductase Enzymes for the Generation of Microbial Production Strains—OptSwap
King ZA, Feist AM. 2013. Optimizing Cofactor Specificity of Oxidoreductase Enzymes for the Generation of Microbial Production Strains—OptSwap. Industrial Biotechnology. 9(4):236-246.
-

The COMBREX project: design, methodology, and initial results.
Anton BP, Chang Y-C, Brown P, Choi H-P, Faller LL, Guleria J, Hu Z, Klitgord N, Levy-Moonshine A, Maksad A et al.. 2013. The COMBREX project: design, methodology, and initial results.. PLoS Biol. 11(8):e1001638.
-

Multispecific Drug Transporter Oat3 (Slc22a8) Regulates Multiple Metabolic Pathways.
Wu W, Jamshidi N, Eraly SA, Liu HC, Bush KT, Palsson BO, Nigam SK. 2013. Multispecific Drug Transporter Oat3 (Slc22a8) Regulates Multiple Metabolic Pathways.. Drug Metab Dispos.
-

Studying Salmonellae and Yersiniae host-pathogen interactions using integrated 'omics and modeling.
Ansong C, Deatherage BL, Hyduke D, Schmidt B, McDermott JE, Jones MB, Chauhan S, Charusanti P, Kim Y-M, Nakayasu ES et al.. 2013. Studying Salmonellae and Yersiniae host-pathogen interactions using integrated 'omics and modeling.. Curr Top Microbiol Immunol. 363:21-41.
-

COBRApy: COnstraints-Based Reconstruction and Analysis for Python.
Ebrahim A, Lerman JA, Palsson BO, Hyduke DR. 2013. COBRApy: COnstraints-Based Reconstruction and Analysis for Python.. BMC Syst Biol. 7(1):74.
-

Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome.
Lewis NE, Liu X, Li Y, Nagarajan H, Yerganian G, O'Brien E, Bordbar A, Roth AM, Rosenbloom J, Bian C et al.. 2013. Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome.. Nat Biotechnol.
-

Structural systems biology evaluation of metabolic thermotolerance in Escherichia coli.
Chang RL, Andrews K, Kim D, Li Z, Godzik A, Palsson BO. 2013. Structural systems biology evaluation of metabolic thermotolerance in Escherichia coli.. Science. 340(6137):1220-3.
-

MS/MS networking guided analysis of molecule and gene cluster families.
Nguyen D D, Wu C-H, Moree WJ, Lamsa A, Medema MH, Zhao X, Gavilan RG, Aparicio M, Atencio L, Jackson C et al.. 2013. MS/MS networking guided analysis of molecule and gene cluster families.. Proc Natl Acad Sci U S A. 110(28):E2611-20.
-

The genome organization of Thermotoga maritima reflects its lifestyle.
Latif H, Lerman JA, Portnoy VA, Tarasova Y, Nagarajan H, Schrimpe-Rutledge AC, Smith RD, Adkins JN, Lee D-H, Qiu Y et al.. 2013. The genome organization of Thermotoga maritima reflects its lifestyle.. PLoS Genet. 9(4):e1003485.
-

Basic and applied uses of genome-scale metabolic network reconstructions of Escherichia coli.
McCloskey D, Palsson BØ, Feist AM. 2013. Basic and applied uses of genome-scale metabolic network reconstructions of Escherichia coli.. Mol Syst Biol. 9:661.
-

Salmonella modulates metabolism during growth under conditions that induce expression of virulence genes.
Kim Y-M, Schmidt BJ, Kidwai AS, Jones MB, Deatherage Kaiser BL, Brewer HM, Mitchell HD, Palsson BO, McDermott JE, Heffron F et al.. 2013. Salmonella modulates metabolism during growth under conditions that induce expression of virulence genes.. Mol Biosyst.
-

Characterizing the interplay between multiple levels of organization within bacterial sigma factor regulatory networks.
Qiu Y, Nagarajan H, Embree M, Shieu W, Abate E, Juárez K, Cho B-K, Elkins JG, Nevin KP, Barrett CL et al.. 2013. Characterizing the interplay between multiple levels of organization within bacterial sigma factor regulatory networks.. Nat Commun. 4:1755.
-

A community-driven global reconstruction of human metabolism.
Thiele I, Swainston N, Fleming RMT, Hoppe A, Sahoo S, Aurich MK, Haraldsdottir H, Mo ML, Rolfsson O, Stobbe MD et al.. 2013. A community-driven global reconstruction of human metabolism.. Nat Biotechnol.
-

The development of a fully-integrated immune response model (FIRM) simulator of the immune response through integration of multiple subset models.
Palsson S, Hickling TP, Bradshaw-Pierce EL, Zager M, Jooss K, Brien PJO, Spilker ME, Palsson BO, Vicini P. 2013. The development of a fully-integrated immune response model (FIRM) simulator of the immune response through integration of multiple subset models.. BMC Syst Biol. 7(1):95.
-

GIM3E: condition-specific models of cellular metabolism developed from metabolomics and expression data.
Schmidt BJ, Ebrahim A, Metz TO, Adkins JN, Palsson BØ, Hyduke DR. 2013. GIM3E: condition-specific models of cellular metabolism developed from metabolomics and expression data.. Bioinformatics.
-

Characterizing acetogenic metabolism using a genome-scale metabolic reconstruction of Clostridium ljungdahlii.
Nagarajan H, Sahin M, Nogales J, Latif H, Lovley DR, Ebrahim A, Zengler K. 2013. Characterizing acetogenic metabolism using a genome-scale metabolic reconstruction of Clostridium ljungdahlii.. Microb Cell Fact. 12(1):118.
-

The microbiome extends to subepidermal compartments of normal skin.
Nakatsuji T, Chiang H-I, Jiang SB, Nagarajan H, Zengler K, Gallo RL. 2013. The microbiome extends to subepidermal compartments of normal skin.. Nat Commun. 4:1431.
-

Reconciling a Salmonella enterica metabolic model with experimental data confirms that overexpression of the glyoxylate shunt can rescue a lethal ppc deletion mutant.
Fong NL, Lerman JA, Lam I, Palsson BO, Charusanti P. 2013. Reconciling a Salmonella enterica metabolic model with experimental data confirms that overexpression of the glyoxylate shunt can rescue a lethal ppc deletion mutant.. FEMS Microbiol Lett. 342(1):62-9.
-

Genomically and biochemically accurate metabolic reconstruction of Methanosarcina barkeri Fusaro, iMG746.
Gonnerman MC, Benedict MN, Feist AM, Metcalf WW, Price ND. 2013. Genomically and biochemically accurate metabolic reconstruction of Methanosarcina barkeri Fusaro, iMG746.. Biotechnol J.
-

Sulfide-driven microbial electrosynthesis.
Gong Y, Ebrahim A, Feist AM, Embree M, Zhang T, Lovley D, Zengler K. 2013. Sulfide-driven microbial electrosynthesis.. Environ Sci Technol. 47(1):568-573.
-

Analysis of omics data with genome-scale models of metabolism.
Hyduke DR, Lewis NE, Palsson BØ. 2013. Analysis of omics data with genome-scale models of metabolism.. Mol Biosyst. 9(2):167-74.
-

Enhancement of carotenoid biosynthesis in the green microalga Dunaliella salina with light-emitting diodes and adaptive laboratory evolution.
Fu W, Guðmundsson O, Paglia G, Herjólfsson G, Andrésson OS, Palsson BO, Brynjólfsson S. 2013. Enhancement of carotenoid biosynthesis in the green microalga Dunaliella salina with light-emitting diodes and adaptive laboratory evolution.. Appl Microbiol Biotechnol. 97(6):2395-403.
-

Inferring the metabolism of human orphan metabolites from their metabolic network context affirms human gluconokinase activity.
Rolfsson O, Paglia G, Magnusdóttir M, Palsson BØ, Thiele I. 2013. Inferring the metabolism of human orphan metabolites from their metabolic network context affirms human gluconokinase activity.. Biochem J. 449(2):427-35.
-
Systems biology of stored blood cells: can it help to extend the expiration date?
Paglia G, Palsson BØ, Sigurjonsson OE. 2012. Systems biology of stored blood cells: can it help to extend the expiration date? J Proteomics. 76 Spec No.:163-7.
-
Proteomic analysis of Chinese hamster ovary cells.
Baycin-Hizal D, Tabb DL, Chaerkady R, Chen L, Lewis NE, Nagarajan H, Sarkaria V, Kumar A, Wolozny D, Colao J et al.. 2012. Proteomic analysis of Chinese hamster ovary cells.. J Proteome Res. 11(11):5265-76.
-
Network context and selection in the evolution to enzyme specificity.
Nam H, Lewis NE, Lerman JA, Lee D-H, Chang RL, Kim D, Palsson BO. 2012. Network context and selection in the evolution to enzyme specificity.. Science. 337(6098):1101-4.
-
Comparative Analysis of Regulatory Elements between Escherichia coli and Klebsiella pneumoniae by Genome-Wide Transcription Start Site Profiling.
Kim D, Hong J S-J, Qiu Y, Nagarajan H, Seo J-H, Cho B-K, Tsai S-F, Palsson BØ. 2012. Comparative Analysis of Regulatory Elements between Escherichia coli and Klebsiella pneumoniae by Genome-Wide Transcription Start Site Profiling.. PLoS Genet. 8(8):e1002867.
-
In silico method for modelling metabolism and gene product expression at genome scale
Lerman JA, Hyduke DR, Latif H, Portnoy VA, Lewis NE, Orth JD, Schrimpe-Rutledge AC, Smith RD, Adkins JN, Zengler K et al.. 2012. In silico method for modelling metabolism and gene product expression at genome scale. Nat Commun. 3:929.
-
Raloxifene attenuates Pseudomonas aeruginosa pyocyanin production and virulence.
Ho Sui SJ, Lo R, Fernandes AR, Caulfield MDG, Lerman JA, Xie L, Bourne PE, Baillie DL, Brinkman FSL. 2012. Raloxifene attenuates Pseudomonas aeruginosa pyocyanin production and virulence.. Int J Antimicrob Agents.
-
Maximizing biomass productivity and cell density of Chlorella vulgaris by using light-emitting diode-based photobioreactor.
Fu W, Gudmundsson O, Feist AM, Herjolfsson G, Brynjolfsson S, Palsson BO. 2012. Maximizing biomass productivity and cell density of Chlorella vulgaris by using light-emitting diode-based photobioreactor.. J Biotechnol.
-
Model-driven multi-omic data analysis elucidates metabolic immunomodulators of macrophage activation.
Bordbar A, Mo ML, Nakayasu ES, Schrimpe-Rutledge AC, Kim Y-M, Metz TO, Jones MB, Frank BC, Smith RD, Peterson SN et al.. 2012. Model-driven multi-omic data analysis elucidates metabolic immunomodulators of macrophage activation.. Mol Syst Biol. 8:558.
-
Gap-filling analysis of the iJO1366 Escherichia coli metabolic network reconstruction for discovery of metabolic functions.
Orth JD, Palsson BO. 2012. Gap-filling analysis of the iJO1366 Escherichia coli metabolic network reconstruction for discovery of metabolic functions.. BMC Syst Biol. 6(1):30.
-
Intracellular metabolite profiling of platelets: evaluation of extraction processes and chromatographic strategies.
Paglia G, Magnúsdóttir M, Thorlacius S, Sigurjónsson OE, Guðmundsson S, Palsson BØ, Thiele I. 2012. Intracellular metabolite profiling of platelets: evaluation of extraction processes and chromatographic strategies.. J Chromatogr B Analyt Technol Biomed Life Sci. 898:111-20.
-
Exploiting adaptive laboratory evolution of Streptomyces clavuligerus for antibiotic discovery and overproduction.
Charusanti P, Fong NL, Nagarajan H, Pereira AR, Li HJ, Abate EA, Su Y, Gerwick WH, Palsson BO. 2012. Exploiting adaptive laboratory evolution of Streptomyces clavuligerus for antibiotic discovery and overproduction.. PLoS One. 7(3):e33727.
-
A road map for the development of community systems (CoSy) biology.
Zengler K, Palsson BO. 2012. A road map for the development of community systems (CoSy) biology.. Nat Rev Microbiol. 10(5):366-72.
-
Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis.
Nogales J, Gudmundsson S, Knight EM, Palsson BO, Thiele I. 2012. Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis.. Proc Natl Acad Sci U S A. 109(7):2678-83.
-
Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods.
Lewis NE, Nagarajan H, Palsson BO. 2012. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods.. Nat Rev Microbiol. 10(4):291-305.
-
Predicting outcomes of steady-state ¹³C isotope tracing experiments using Monte Carlo sampling.
Schellenberger J, Zielinski DC, Choi W, Madireddi S, Portnoy V, Scott DA, Reed JL, Osterman AL, Palsson B. 2012. Predicting outcomes of steady-state ¹³C isotope tracing experiments using Monte Carlo sampling.. BMC Syst Biol. 6:9.
-
Using the reconstructed genome-scale human metabolic network to study physiology and pathology.
Bordbar A, Palsson BO. 2012. Using the reconstructed genome-scale human metabolic network to study physiology and pathology.. J Intern Med. 271(2):131-41.
-
MODELING HOST-PATHOGEN INTERACTIONS: COMPUTATIONAL BIOLOGY AND BIOINFORMATICS FOR INFECTIOUS DISEASE RESEARCH.
McDermott JE, Braun P, Bonneau R, Hyduke DR. 2012. MODELING HOST-PATHOGEN INTERACTIONS: COMPUTATIONAL BIOLOGY AND BIOINFORMATICS FOR INFECTIOUS DISEASE RESEARCH.. Pac Symp Biocomput. 17:283-286.
-
Monitoring metabolites consumption and secretion in cultured cells using ultra-performance liquid chromatography quadrupole-time of flight mass spectrometry (UPLC-Q-ToF-MS).
Paglia G, Hrafnsdóttir S, Magnúsdóttir M, Fleming RMT, Thorlacius S, Palsson BØ, Thiele I. 2012. Monitoring metabolites consumption and secretion in cultured cells using ultra-performance liquid chromatography quadrupole-time of flight mass spectrometry (UPLC-Q-ToF-MS).. Anal Bioanal Chem. 402(3):1183-98.
-
Multiple-omic data analysis of Klebsiella pneumoniae MGH 78578 reveals its transcriptional architecture and regulatory features.
Seo J-H, Hong J S-J, Kim D, Cho B-K, Huang T-W, Tsai S-F, Palsson BO, Charusanti P. 2012. Multiple-omic data analysis of Klebsiella pneumoniae MGH 78578 reveals its transcriptional architecture and regulatory features.. BMC Genomics. 13(1):679.
-
Anaerobic utilization of toluene by marine alpha- and gammaproteobacteria reducing nitrate.
Alain K, Harder J, Widdel F, Zengler K. 2012. Anaerobic utilization of toluene by marine alpha- and gammaproteobacteria reducing nitrate.. Microbiology. 158(Pt 12):2946-57.
-
Multiscale Modeling of Metabolism and Macromolecular Synthesis in E. coli and Its Application to the Evolution of Codon Usage.
Thiele I, Fleming RMT, Que R, Bordbar A, Diep D, Palsson BO. 2012. Multiscale Modeling of Metabolism and Macromolecular Synthesis in E. coli and Its Application to the Evolution of Codon Usage.. PLoS One. 7(9):e45635.
-
UPLC-UV-MS(E) analysis for quantification and identification of major carotenoid and chlorophyll species in algae.
Fu W, Magnúsdóttir M, Brynjólfson S, Palsson BO, Paglia G. 2012. UPLC-UV-MS(E) analysis for quantification and identification of major carotenoid and chlorophyll species in algae.. Anal Bioanal Chem. 404(10):3145-54.
-
A variational principle for computing nonequilibrium fluxes and potentials in genome-scale biochemical networks.
Fleming RMT, Maes CM, Saunders MA, Ye Y, Palsson BØ. 2012. A variational principle for computing nonequilibrium fluxes and potentials in genome-scale biochemical networks.. J Theor Biol. 292:71-7.
-
Transcriptional regulation of central carbon and energy metabolism in bacteria by redox-responsive repressor Rex.
Ravcheev DA, Li X, Latif H, Zengler K, Leyn SA, Korostelev YD, Kazakov AE, Novichkov PS, Osterman AL, Rodionov DA. 2012. Transcriptional regulation of central carbon and energy metabolism in bacteria by redox-responsive repressor Rex.. J Bacteriol. 194(5):1145-57.
-
Deciphering the transcriptional regulatory logic of amino acid metabolism.
Cho B-K, Federowicz S, Park Y-S, Zengler K, Palsson BØ. 2011. Deciphering the transcriptional regulatory logic of amino acid metabolism.. Nat Chem Biol. 8(1):65-71.
-
Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0.
Schellenberger J, Que R, Fleming RMT, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S et al.. 2011. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0.. Nat Protoc. 6(9):1290-307.
-
Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism.
Chang RL, Ghamsari L, Manichaikul A, Hom EFY, Balaji S, Fu W, Shen Y, Hao T, Palsson BØ, Salehi-Ashtiani K et al.. 2011. Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism.. Mol Syst Biol. 7:518.
-
The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line.
Xu X, Nagarajan H, Lewis NE, Pan S, Cai Z, Liu X, Chen W, Xie M, Wang W, Hammond S et al.. 2011. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line.. Nat Biotechnol. 29(8):735-41.
-
Microbial laboratory evolution in the era of genome-scale science.
Conrad TM, Lewis NE, Palsson BØ. 2011. Microbial laboratory evolution in the era of genome-scale science.. Mol Syst Biol. 7:509.
-
A c-type cytochrome and a transcriptional regulator responsible for enhanced extracellular electron transfer in Geobacter sulfurreducens revealed by adaptive evolution.
Tremblay P-L, Summers ZM, Glaven RH, Nevin KP, Zengler K, Barrett CL, Qiu Y, Palsson BO, Lovley DR. 2011. A c-type cytochrome and a transcriptional regulator responsible for enhanced extracellular electron transfer in Geobacter sulfurreducens revealed by adaptive evolution.. Environ Microbiol. 13(1):13-23.
-
Linkage of organic anion transporter-1 to metabolic pathways through integrated "omics"-driven network and functional analysis.
Ahn S-Y, Jamshidi N, Mo ML, Wu W, Eraly SA, Dnyanmote A, Bush KT, Gallegos TF, Sweet DH, Palsson BØ et al.. 2011. Linkage of organic anion transporter-1 to metabolic pathways through integrated "omics"-driven network and functional analysis.. J Biol Chem. 286(36):31522-31.
-
iAB-RBC-283: A proteomically derived knowledge-base of erythrocyte metabolism that can be used to simulate its physiological and patho-physiological states.
Bordbar A, Jamshidi N, Palsson BO. 2011. iAB-RBC-283: A proteomically derived knowledge-base of erythrocyte metabolism that can be used to simulate its physiological and patho-physiological states.. BMC Syst Biol. 5:110.
-
Technologies and approaches to elucidate and model the virulence program of salmonella.
McDermott JE, Yoon H, Nakayasu ES, Metz TO, Hyduke DR, Kidwai AS, Palsson BO, Adkins JN, Heffron F. 2011. Technologies and approaches to elucidate and model the virulence program of salmonella.. Front Microbiol. 2:121.
-
The PurR regulon in Escherichia coli K-12 MG1655.
Cho B-K, Federowicz SA, Embree M, Park Y-S, Kim D, Palsson BØ. 2011. The PurR regulon in Escherichia coli K-12 MG1655.. Nucleic Acids Res. 39(15):6456-64.
-
Functional and metabolic effects of adaptive glycerol kinase (GLPK) mutants in Escherichia coli.
Applebee KM, Joyce AR, Conrad TM, Pettigrew DW, Palsson BØ. 2011. Functional and metabolic effects of adaptive glycerol kinase (GLPK) mutants in Escherichia coli.. J Biol Chem. 286(26):23150-9.
-
Adaptive laboratory evolution--harnessing the power of biology for metabolic engineering.
Portnoy VA, Bezdan D, Zengler K. 2011. Adaptive laboratory evolution--harnessing the power of biology for metabolic engineering.. Curr Opin Biotechnol. 22(4):590-4.
-
The role of cellular objectives and selective pressures in metabolic pathway evolution.
Nam H, Conrad TM, Lewis NE. 2011. The role of cellular objectives and selective pressures in metabolic pathway evolution.. Curr Opin Biotechnol. 22(4):595-600.
-
An experimentally validated genome-scale metabolic reconstruction of Klebsiella pneumoniae MGH 78578, iYL1228.
Liao Y-C, Huang T-W, Chen F-C, Charusanti P, Hong JSJ, Chang H-Y, Tsai S-F, Palsson BO, Hsiung CA. 2011. An experimentally validated genome-scale metabolic reconstruction of Klebsiella pneumoniae MGH 78578, iYL1228.. J Bacteriol. 193(7):1710-7.
-
Elimination of thermodynamically infeasible loops in steady-state metabolic models.
Schellenberger J, Lewis NE, Palsson BØ. 2011. Elimination of thermodynamically infeasible loops in steady-state metabolic models.. Biophys J. 100(3):544-53.
-
-
In situ to in silico and back: elucidating the physiology and ecology of Geobacter spp. using genome-scale modelling.
Mahadevan R, Palsson BØ, Lovley DR. 2011. In situ to in silico and back: elucidating the physiology and ecology of Geobacter spp. using genome-scale modelling.. Nat Rev Microbiol. 9(1):39-50.
-
Sensitive and accurate identification of protein-DNA binding events in ChIP-chip assays using higher order derivative analysis.
Barrett CL, Cho B-K, Palsson BO. 2011. Sensitive and accurate identification of protein-DNA binding events in ChIP-chip assays using higher order derivative analysis.. Nucleic Acids Res. 39(5):1656-65.
-
A multi-tissue type genome-scale metabolic network for analysis of whole-body systems physiology.
Bordbar A, Feist AM, Usaite-Black R, Woodcock J, Palsson BO, Famili I. 2011. A multi-tissue type genome-scale metabolic network for analysis of whole-body systems physiology.. BMC Syst Biol. 5:180.
-
The human metabolic reconstruction Recon 1 directs hypotheses of novel human metabolic functions.
Rolfsson O, Palsson BØ, Thiele I. 2011. The human metabolic reconstruction Recon 1 directs hypotheses of novel human metabolic functions.. BMC Syst Biol. 5:155.
-
Cumulative number of cell divisions as a meaningful timescale for adaptive laboratory evolution of Escherichia coli.
Lee D-H, Feist AM, Barrett CL, Palsson BØ. 2011. Cumulative number of cell divisions as a meaningful timescale for adaptive laboratory evolution of Escherichia coli.. PLoS One. 6(10):e26172.
-
An experimentally-supported genome-scale metabolic network reconstruction for Yersinia pestis CO92.
Charusanti P, Chauhan S, McAteer K, Lerman JA, Hyduke DR, Motin VL, Ansong C, Adkins JN, Palsson BO. 2011. An experimentally-supported genome-scale metabolic network reconstruction for Yersinia pestis CO92.. BMC Syst Biol. 5:163.
-
A comprehensive genome-scale reconstruction of Escherichia coli metabolism--2011.
Orth JD, Conrad TM, Na J, Lerman JA, Nam H, Feist AM, Palsson BØ. 2011. A comprehensive genome-scale reconstruction of Escherichia coli metabolism--2011.. Mol Syst Biol. 7:535.
-
A community effort towards a knowledge-base and mathematical model of the human pathogen Salmonella Typhimurium LT2.
Thiele I, Hyduke DR, Steeb B, Fankam G, Allen DK, Bazzani S, Charusanti P, Chen F-C, Fleming RMT, Hsiung CA et al.. 2011. A community effort towards a knowledge-base and mathematical model of the human pathogen Salmonella Typhimurium LT2.. BMC Syst Biol. 5:8.
-
Production of pilus-like filaments in Geobacter sulfurreducens in the absence of the type IV pilin protein PilA.
Klimes A, Franks AE, Glaven RH, Tran H, Barrett CL, Qiu Y, Zengler K, Lovley DR. 2010. Production of pilus-like filaments in Geobacter sulfurreducens in the absence of the type IV pilin protein PilA.. FEMS microbiology letters. 310(1):62-8.PubMed Google Scholar
-
Structural and operational complexity of the Geobacter sulfurreducens genome.
Qiu Y, Cho B-K, Park Y S, Lovley DR, Palsson BØ, Zengler K. 2010. Structural and operational complexity of the Geobacter sulfurreducens genome.. Genome research. 20(9):1304-11.PubMed Google Scholar
-
Insight into human alveolar macrophage and M. tuberculosis interactions via metabolic reconstructions.
Bordbar A, Lewis NE, Schellenberger J, Palsson BØ, Jamshidi N. 2010. Insight into human alveolar macrophage and M. tuberculosis interactions via metabolic reconstructions.. Molecular systems biology. 6:422.
-
Systematizing the generation of missing metabolic knowledge.
Orth JD, Palsson BØ. 2010. Systematizing the generation of missing metabolic knowledge.. Biotechnology and bioengineering. 107(3):403-12.
-
Deletion of genes encoding cytochrome oxidases and quinol monooxygenase blocks the aerobic-anaerobic shift in Escherichia coli K-12 MG1655.
Portnoy VA, Scott DA, Lewis NE, Tarasova Y, Osterman AL, Palsson BØ. 2010. Deletion of genes encoding cytochrome oxidases and quinol monooxygenase blocks the aerobic-anaerobic shift in Escherichia coli K-12 MG1655.. Applied and environmental microbiology. 76(19):6529-40.
-
RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media.
Conrad TM, Frazier M, Joyce AR, Cho B-K, Knight EM, Lewis NE, Landick R, Palsson BØ. 2010. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media.. Proceedings of the National Academy of Sciences of the United States of America. 107(47):20500-5.
-
Microbiology. Topping off a multiscale balancing act.
Lerman JA, Palsson BØ. 2010. Microbiology. Topping off a multiscale balancing act.. Science (New York, N.Y.). 330(6007):1058-9.
-
The challenges of integrating multi-omic data sets.
Palsson BØ, Zengler K. 2010. The challenges of integrating multi-omic data sets.. Nature chemical biology. 6(11):787-9.
-
Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene.
Charusanti P, Conrad TM, Knight EM, Venkataraman K, Fong NL, Xie B, Gao Y, Palsson BØ. 2010. Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene.. PLoS genetics. 6(11):e1001186.
-
Functional characterization of alternate optimal solutions of Escherichia coli's transcriptional and translational machinery.
Thiele I, Fleming RMT, Bordbar A, Schellenberger J, Palsson BØ. 2010. Functional characterization of alternate optimal solutions of Escherichia coli's transcriptional and translational machinery.. Biophysical journal. 98(10):2072-81.
-
Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli.
Feist AM, Zielinski DC, Orth JD, Schellenberger J, Herrgard MJ, Palsson BØ. 2010. Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli.. Metabolic engineering. 12(3):173-86.
-
What is flux balance analysis?
Orth JD, Thiele I, Palsson BØ. 2010. What is flux balance analysis? Nature biotechnology. 28(3):245-8.
-
The biomass objective function.
Feist AM, Palsson BØ. 2010. The biomass objective function.. Current opinion in microbiology. 13(3):344-9.
-
Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a Nonnative carbon source, L-1,2-propanediol.
Lee D-H, Palsson BØ. 2010. Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a Nonnative carbon source, L-1,2-propanediol.. Applied and environmental microbiology. 76(13):4158-68.
-
Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models.
Lewis NE, Hixson KK, Conrad TM, Lerman JA, Charusanti P, Polpitiya AD, Adkins JN, Schramm G, Purvine SO, Lopez-Ferrer D et al.. 2010. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models.. Molecular systems biology. 6:390.
-
Mass action stoichiometric simulation models: incorporating kinetics and regulation into stoichiometric models.
Jamshidi N, Palsson BØ. 2010. Mass action stoichiometric simulation models: incorporating kinetics and regulation into stoichiometric models.. Biophysical journal. 98(2):175-85.
-
Large-scale in silico modeling of metabolic interactions between cell types in the human brain.
Lewis NE, Schramm G, Bordbar A, Schellenberger J, Andersen MP, Cheng JK, Patel N, Yee A, Lewis RA, Eils R et al.. 2010. Large-scale in silico modeling of metabolic interactions between cell types in the human brain.. Nature biotechnology. 28(12):1279-85.
-
Reconstruction annotation jamborees: a community approach to systems biology.
Thiele I, Palsson BØ. 2010. Reconstruction annotation jamborees: a community approach to systems biology.. Molecular systems biology. 6:361.
-
Towards genome-scale signalling network reconstructions.
Hyduke DR, Palsson BØ. 2010. Towards genome-scale signalling network reconstructions.. Nature reviews. Genetics. 11(4):297-307.
-
BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions.
Schellenberger J, Park JO, Conrad TM, Palsson BØ. 2010. BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions.. BMC bioinformatics. 11:213.
-
De Novo assembly of the complete genome of an enhanced electricity-producing variant of Geobacter sulfurreducens using only short reads.
Nagarajan H, Butler JE, Klimes A, Qiu Y, Zengler K, Ward J, Young ND, Methé BA, Palsson BØ, Lovley DR et al.. 2010. De Novo assembly of the complete genome of an enhanced electricity-producing variant of Geobacter sulfurreducens using only short reads.. PloS one. 5(6):e10922.
-
Drug off-target effects predicted using structural analysis in the context of a metabolic network model.
Chang RL, Xie L, Xie L, Bourne PE, Palsson BØ. 2010. Drug off-target effects predicted using structural analysis in the context of a metabolic network model.. PLoS computational biology. 6(9):e1000938.
-
A detailed genome-wide reconstruction of mouse metabolism based on human Recon 1.
Sigurdsson MI, Jamshidi N, Steingrimsson E, Thiele I, Palsson BØ. 2010. A detailed genome-wide reconstruction of mouse metabolism based on human Recon 1.. BMC systems biology. 4:140.
-
A protocol for generating a high-quality genome-scale metabolic reconstruction.
Thiele I, Palsson BØ. 2010. A protocol for generating a high-quality genome-scale metabolic reconstruction.. Nature protocols. 5(1):93-121.
-
Flux-concentration duality in dynamic nonequilibrium biological networks.
Jamshidi N, Palsson BØ. 2009. Flux-concentration duality in dynamic nonequilibrium biological networks.. Biophysical journal. 97(5):L11-3.
-
Three-dimensional structural view of the central metabolic network of Thermotoga maritima.
Zhang Y, Thiele I, Weekes D, Li Z, Jaroszewski L, Ginalski K, Deacon AM, Wooley J, Lesley SA, Wilson IA et al.. 2009. Three-dimensional structural view of the central metabolic network of Thermotoga maritima.. Science (New York, N.Y.). 325(5947):1544-9.
-
The transcription unit architecture of the Escherichia coli genome.
Cho B-K, Zengler K, Qiu Y, Park Y S, Knight EM, Barrett CL, Gao Y, Palsson BØ. 2009. The transcription unit architecture of the Escherichia coli genome.. Nature biotechnology. 27(11):1043-9.
-
Genome-scale reconstruction of Escherichia coli's transcriptional and translational machinery: a knowledge base, its mathematical formulation, and its functional characterization.
Thiele I, Jamshidi N, Fleming RMT, Palsson BØ. 2009. Genome-scale reconstruction of Escherichia coli's transcriptional and translational machinery: a knowledge base, its mathematical formulation, and its functional characterization.. PLoS computational biology. 5(3):e1000312.
-
Gene expression profiling and the use of genome-scale in silico models of Escherichia coli for analysis: providing context for content.
Lewis NE, Cho B-K, Knight EM, Palsson BØ. 2009. Gene expression profiling and the use of genome-scale in silico models of Escherichia coli for analysis: providing context for content.. Journal of bacteriology. 191(11):3437-44.
-
Functional states of the genome-scale Escherichia coli transcriptional regulatory system.
Gianchandani EP, Joyce AR, Palsson BØ, Papin JA. 2009. Functional states of the genome-scale Escherichia coli transcriptional regulatory system.. PLoS computational biology. 5(6):e1000403.
-
Genome-scale network analysis of imprinted human metabolic genes.
Sigurdsson MI, Jamshidi N, Jonsson JJ, Palsson BØ. 2009. Genome-scale network analysis of imprinted human metabolic genes.. Epigenetics : official journal of the DNA Methylation Society. 4(1):43-6.
-
Understanding human metabolic physiology: a genome-to-systems approach.
Mo ML, Palsson BØ. 2009. Understanding human metabolic physiology: a genome-to-systems approach.. Trends in biotechnology. 27(1):37-44.
-
Use of randomized sampling for analysis of metabolic networks.
Schellenberger J, Palsson BØ. 2009. Use of randomized sampling for analysis of metabolic networks.. The Journal of biological chemistry. 284(9):5457-61.
-
Reconstruction of biochemical networks in microorganisms.
Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BØ. 2009. Reconstruction of biochemical networks in microorganisms.. Nature reviews. Microbiology. 7(2):129-43.
-
Identification of potential pathway mediation targets in Toll-like receptor signaling.
Li F, Thiele I, Jamshidi N, Palsson BØ. 2009. Identification of potential pathway mediation targets in Toll-like receptor signaling.. PLoS computational biology. 5(2):e1000292.
-
Metabolic systems biology.
Palsson BØ. 2009. Metabolic systems biology.. FEBS letters. 583(24):3900-4.
-
Can the protein occupancy landscape show the topologically isolated chromosomal domains in the E. coli genome?: An exciting prospect.
Cho B-K, Palsson BØ. 2009. Can the protein occupancy landscape show the topologically isolated chromosomal domains in the E. coli genome?: An exciting prospect. Molecular cell. 35(3):255-6.
-
Metabolic network analysis integrated with transcript verification for sequenced genomes.
Manichaikul A, Ghamsari L, Hom EFY, Lin C, Murray RR, Chang RL, Balaji S, Hao T, Shen Y, Chavali AK et al.. 2009. Metabolic network analysis integrated with transcript verification for sequenced genomes.. Nature methods. 6(8):589-92.
-
Probing the basis for genotype-phenotype relationships.
Cho B-K, Palsson BØ. 2009. Probing the basis for genotype-phenotype relationships.. Nature methods. 6(8):565-6.
-
Constraint-based analysis of metabolic capacity of Salmonella typhimurium during host-pathogen interaction.
Raghunathan A, Reed JL, Shin S, Palsson BØ, Daefler S. 2009. Constraint-based analysis of metabolic capacity of Salmonella typhimurium during host-pathogen interaction.. BMC systems biology. 3:38.
-
Connecting extracellular metabolomic measurements to intracellular flux states in yeast.
Mo ML, Palsson BØ, Herrgard MJ. 2009. Connecting extracellular metabolomic measurements to intracellular flux states in yeast.. BMC systems biology. 3:37.
-
Using in silico models to simulate dual perturbation experiments: procedure development and interpretation of outcomes.
Jamshidi N, Palsson BØ. 2009. Using in silico models to simulate dual perturbation experiments: procedure development and interpretation of outcomes.. BMC systems biology. 3:44.
-
Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations.
Conrad TM, Joyce AR, Applebee KM, Barrett CL, Xie B, Gao Y, Palsson BØ. 2009. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations.. Genome biology. 10(10):R118.
-
Applications of genome-scale metabolic reconstructions.
Oberhardt MA, Palsson BØ, Papin JA. 2009. Applications of genome-scale metabolic reconstructions.. Molecular systems biology. 5:320.
-
Decomposing complex reaction networks using random sampling, principal component analysis and basis rotation.
Barrett CL, Herrgard MJ, Palsson BØ. 2009. Decomposing complex reaction networks using random sampling, principal component analysis and basis rotation.. BMC systems biology. 3:30.
-
Network-based prediction of human tissue-specific metabolism.
Shlomi T, Cabili MN, Herrgard MJ, Palsson BØ, Ruppin E. 2008. Network-based prediction of human tissue-specific metabolism.. Nature biotechnology. 26(9):1003-10.
-
Genome-scale reconstruction and in silico analysis of the Clostridium acetobutylicum ATCC 824 metabolic network.
Lee J, Yun H, Feist AM, Palsson BØ, Lee S Y. 2008. Genome-scale reconstruction and in silico analysis of the Clostridium acetobutylicum ATCC 824 metabolic network.. Applied microbiology and biotechnology. 80(5):849-62.
-
A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology.
Herrgard MJ, Swainston N, Dobson P, Dunn WB, Arga YK, Arvas M, Blüthgen N, Borger S, Costenoble R, Heinemann M et al.. 2008. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology.. Nature biotechnology. 26(10):1155-60.
-
Context-specific metabolic networks are consistent with experiments.
Becker SA, Palsson BØ. 2008. Context-specific metabolic networks are consistent with experiments.. PLoS computational biology. 4(5):e1000082.PubMed Google Scholar
-
Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts.
Cho B-K, Knight EM, Barrett CL, Palsson BØ. 2008. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts.. Genome research. 18(6):900-10.
-
The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli.
Feist AM, Palsson BØ. 2008. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli.. Nature biotechnology. 26(6):659-67.
-
Impact of individual mutations on increased fitness in adaptively evolved strains of Escherichia coli.
Applebee KM, Herrgard MJ, Palsson BØ. 2008. Impact of individual mutations on increased fitness in adaptively evolved strains of Escherichia coli.. Journal of bacteriology. 190(14):5087-94.
-
Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli.
Cho B-K, Barrett CL, Knight EM, Park Y S, Palsson BØ. 2008. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli.. Proceedings of the National Academy of Sciences of the United States of America. 105(49):19462-7.
-
Aerobic fermentation of D-glucose by an evolved cytochrome oxidase-deficient Escherichia coli strain.
Portnoy VA, Herrgard MJ, Palsson BØ. 2008. Aerobic fermentation of D-glucose by an evolved cytochrome oxidase-deficient Escherichia coli strain.. Applied and environmental microbiology. 74(24):7561-9.
-
Formulating genome-scale kinetic models in the post-genome era.
Jamshidi N, Palsson BØ. 2008. Formulating genome-scale kinetic models in the post-genome era.. Molecular systems biology. 4:171.
-
Genomewide identification of protein binding locations using chromatin immunoprecipitation coupled with microarray.
Cho B-K, Knight EM, Palsson BØ. 2008. Genomewide identification of protein binding locations using chromatin immunoprecipitation coupled with microarray.. Methods in molecular biology (Clifton, N.J.). 439:131-45.
-
Predicting gene essentiality using genome-scale in silico models.
Joyce AR, Palsson BØ. 2008. Predicting gene essentiality using genome-scale in silico models.. Methods in molecular biology (Clifton, N.J.). 416:433-57.
-
Top-down analysis of temporal hierarchy in biochemical reaction networks.
Jamshidi N, Palsson BØ. 2008. Top-down analysis of temporal hierarchy in biochemical reaction networks.. PLoS computational biology. 4(9):e1000177.
-
A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory.
Nogales J, Palsson BØ, Thiele I. 2008. A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory.. BMC systems biology. 2:79.
-
Three factors underlying incorrect in silico predictions of essential metabolic genes.
Becker SA, Palsson BØ. 2008. Three factors underlying incorrect in silico predictions of essential metabolic genes.. BMC systems biology. 2:14.
-
Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data.
Oh Y-K, Palsson BØ, Park SM, Schilling CH, Mahadevan R. 2007. Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data.. The Journal of biological chemistry. 282(39):28791-9.
-
A genome-scale, constraint-based approach to systems biology of human metabolism.
Mo ML, Jamshidi N, Palsson BØ. 2007. A genome-scale, constraint-based approach to systems biology of human metabolism.. Molecular bioSystems. 3(9):598-603.
-
Metabolic reconstruction and modeling of nitrogen fixation in Rhizobium etli.
Resendis-Antonio O, Reed JL, Encarnación S, Collado-Vides J, Palsson BØ. 2007. Metabolic reconstruction and modeling of nitrogen fixation in Rhizobium etli.. PLoS computational biology. 3(10):1887-95.
-
Systematic condition-dependent annotation of metabolic genes.
Shlomi T, Herrgard MJ, Portnoy VA, Naim E, Palsson BØ, Sharan R, Ruppin E. 2007. Systematic condition-dependent annotation of metabolic genes.. Genome research. 17(11):1626-33.
-
Crystal structure of a hyperactive Escherichia coli glycerol kinase mutant Gly230 --> Asp obtained using microfluidic crystallization devices.
Anderson MJ, DeLabarre B, Raghunathan A, Palsson BØ, Brunger AT, Quake SR. 2007. Crystal structure of a hyperactive Escherichia coli glycerol kinase mutant Gly230 --> Asp obtained using microfluidic crystallization devices.. Biochemistry. 46(19):5722-31.
-
Systems analysis of energy metabolism elucidates the affected respiratory chain complex in Leigh's syndrome.
Vo TD, Paul Lee WN, Palsson BØ. 2007. Systems analysis of energy metabolism elucidates the affected respiratory chain complex in Leigh's syndrome.. Molecular genetics and metabolism. 91(1):15-22.
-
Metabolic characterization of Escherichia coli strains adapted to growth on lactate.
Hua Q, Joyce AR, Palsson BØ, Fong S. 2007. Metabolic characterization of Escherichia coli strains adapted to growth on lactate.. Applied and environmental microbiology. 73(14):4639-47.
-
Building the power house: recent advances in mitochondrial studies through proteomics and systems biology.
Vo TD, Palsson BØ. 2007. Building the power house: recent advances in mitochondrial studies through proteomics and systems biology.. American journal of physiology. Cell physiology. 292(1):C164-77.
-
Global reconstruction of the human metabolic network based on genomic and bibliomic data.
Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BØ. 2007. Global reconstruction of the human metabolic network based on genomic and bibliomic data.. Proceedings of the National Academy of Sciences of the United States of America. 104(6):1777-82.
-
Microbial regulatory and metabolic networks.
Cho B-K, Charusanti P, Herrgard MJ, Palsson BØ. 2007. Microbial regulatory and metabolic networks.. Current opinion in biotechnology. 18(4):360-4.
-
Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox.
Becker SA, Feist AM, Mo ML, Hannum G, Palsson BØ, Herrgard MJ. 2007. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox.. Nature protocols. 2(3):727-38.
-
Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets.
Jamshidi N, Palsson BØ. 2007. Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets.. BMC systems biology. 1:26.
-
In silico analysis of SNPs and other high-throughput data.
Jamshidi N, Vo TD, Palsson BØ. 2007. In silico analysis of SNPs and other high-throughput data.. Methods in molecular biology (Clifton, N.J.). 366:267-85.
-
A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information.
Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BØ. 2007. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information.. Molecular systems biology. 3:121.
-
An evaluation of Comparative Genome Sequencing (CGS) by comparing two previously-sequenced bacterial genomes.
Herring CD, Palsson BØ. 2007. An evaluation of Comparative Genome Sequencing (CGS) by comparing two previously-sequenced bacterial genomes.. BMC genomics. 8:274.
-
Estimation of the number of extreme pathways for metabolic networks.
Yeung M, Thiele I, Palsson BØ. 2007. Estimation of the number of extreme pathways for metabolic networks.. BMC bioinformatics. 8(1):363.
-
Toward whole cell modeling and simulation: comprehensive functional genomics through the constraint-based approach.
Joyce AR, Palsson BØ. 2007. Toward whole cell modeling and simulation: comprehensive functional genomics through the constraint-based approach.. Progress in drug research. Fortschritte der Arzneimittelforschung. Progrès des recherches pharmaceutiques. 64:265,267-309.
-
Systems biology as a foundation for genome-scale synthetic biology.
Barrett CL, Kim T Y, Kim H U, Palsson BØ, Lee S Y. 2006. Systems biology as a foundation for genome-scale synthetic biology.. Current opinion in biotechnology. 17(5):488-92.
-
Systems approach to refining genome annotation.
Reed JL, Patel TR, Chen KH, Joyce AR, Applebee KM, Herring CD, Bui OT, Knight EM, Fong S, Palsson BØ. 2006. Systems approach to refining genome annotation.. Proceedings of the National Academy of Sciences of the United States of America. 103(46):17480-4.
-
Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae.
Herrgard MJ, Lee B-S, Portnoy VA, Palsson BØ. 2006. Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae.. Genome research. 16(5):627-35.
-
Iterative reconstruction of transcriptional regulatory networks: an algorithmic approach.
Barrett CL, Palsson BØ. 2006. Iterative reconstruction of transcriptional regulatory networks: an algorithmic approach.. PLoS computational biology. 2(5):e52.
-
Systems biology of the human red blood cell.
Jamshidi N, Palsson BØ. 2006. Systems biology of the human red blood cell.. Blood cells, molecules & diseases. 36(2):239-47.
-
Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes.
Fong S, Nanchen A, Palsson BØ, Sauer U. 2006. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes.. The Journal of biological chemistry. 281(12):8024-33.
-
The model organism as a system: integrating 'omics' data sets.
Joyce AR, Palsson BØ. 2006. The model organism as a system: integrating 'omics' data sets.. Nature reviews. Molecular cell biology. 7(3):198-210.
-
Candidate states of Helicobacter pylori's genome-scale metabolic network upon application of "loop law" thermodynamic constraints.
Price ND, Thiele I, Palsson BØ. 2006. Candidate states of Helicobacter pylori's genome-scale metabolic network upon application of "loop law" thermodynamic constraints.. Biophysical journal. 90(11):3919-28.
-
Identification of genome-scale metabolic network models using experimentally measured flux profiles.
Herrgard MJ, Fong S, Palsson BØ. 2006. Identification of genome-scale metabolic network models using experimentally measured flux profiles.. PLoS computational biology. 2(7):e72.r
-
Optoinjection for efficient targeted delivery of a broad range of compounds and macromolecules into diverse cell types.
Clark IB, Hanania EG, Stevens J, Gallina M, Fieck A, Brandes R, Palsson BØ, Koller MR. 2006. Optoinjection for efficient targeted delivery of a broad range of compounds and macromolecules into diverse cell types.. Journal of biomedical optics. 11(1):014034.
-
Long-range periodic patterns in microbial genomes indicate significant multi-scale chromosomal organization.
Allen TE, Price ND, Joyce AR, Palsson BØ. 2006. Long-range periodic patterns in microbial genomes indicate significant multi-scale chromosomal organization.. PLoS computational biology. 2(1):e2.
-
PCR-based tandem epitope tagging system for Escherichia coli genome engineering.
Cho B-K, Knight EM, Palsson BØ. 2006. PCR-based tandem epitope tagging system for Escherichia coli genome engineering.. BioTechniques. 40(1):67-72.
-
Towards multidimensional genome annotation.
Reed JL, Famili I, Thiele I, Palsson BØ. 2006. Towards multidimensional genome annotation.. Nature reviews. Genetics. 7(2):130-41.
-
Characterization of metabolism in the Fe(III)-reducing organism Geobacter sulfurreducens by constraint-based modeling.
Mahadevan R, Bond DR, Butler JE, Esteve-Nuñez A, Coppi MV, Palsson BØ, Schilling CH, Lovley DR. 2006. Characterization of metabolism in the Fe(III)-reducing organism Geobacter sulfurreducens by constraint-based modeling.. Applied and environmental microbiology. 72(2):1558-68.
-
Metabolic analysis of adaptive evolution for in silico-designed lactate-producing strains.
Hua Q, Joyce AR, Fong S, Palsson BØ. 2006. Metabolic analysis of adaptive evolution for in silico-designed lactate-producing strains.. Biotechnology and bioengineering. 95(5):992-1002.
-
Isotopomer analysis of myocardial substrate metabolism: a systems biology approach.
Vo TD, Palsson BØ. 2006. Isotopomer analysis of myocardial substrate metabolism: a systems biology approach.. Biotechnology and bioengineering. 95(5):972-83.
-
Experimental and computational assessment of conditionally essential genes in Escherichia coli.
Joyce AR, Reed JL, White A, Edwards R, Osterman AL, Baba T, Mori H, Lesely SA, Palsson BØ, Agarwalla S. 2006. Experimental and computational assessment of conditionally essential genes in Escherichia coli.. Journal of bacteriology. 188(23):8259-71.
-
Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale.
Herring CD, Raghunathan A, Honisch C, Patel T, Applebee KM, Joyce AR, Albert TJ, Blattner FR, van den Boom D, Cantor CR et al.. 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale.. Nature genetics. 38(12):1406-12.
-
Matrix formalism to describe functional states of transcriptional regulatory systems.
Gianchandani EP, Papin JA, Price ND, Joyce AR, Palsson BØ. 2006. Matrix formalism to describe functional states of transcriptional regulatory systems.. PLoS computational biology. 2(8):e101.
-
Transcriptional regulation of the fad regulon genes of Escherichia coli by ArcA.
Cho B-K, Knight EM, Palsson BØ. 2006. Transcriptional regulation of the fad regulon genes of Escherichia coli by ArcA.. Microbiology (Reading, England). 152(Pt 8):2207-19.
-
Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri.
Feist AM, Scholten JCM, Palsson BØ, Brockman FJ, Ideker T. 2006. Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri.. Molecular systems biology. 2:2006.0004.
-
Systems biology of SNPs.
Jamshidi N, Palsson BØ. 2006. Systems biology of SNPs.. Molecular systems biology. 2:38.
-
Metabolite coupling in genome-scale metabolic networks.
Becker SA, Price ND, Palsson BØ. 2006. Metabolite coupling in genome-scale metabolic networks.. BMC bioinformatics. 7:111.
-
Network-level analysis of metabolic regulation in the human red blood cell using random sampling and singular value decomposition.
Barrett CL, Price ND, Palsson BØ. 2006. Network-level analysis of metabolic regulation in the human red blood cell using random sampling and singular value decomposition.. BMC bioinformatics. 7:132.
-
In silico design and adaptive evolution of Escherichia coli for production of lactic acid.
Fong S, Burgard AP, Herring CD, Knight EM, Blattner FR, Maranas CD, Palsson BØ. 2005. In silico design and adaptive evolution of Escherichia coli for production of lactic acid.. Biotechnology and bioengineering. 91(5):643-8.
-
Automated in situ measurement of cell-specific antibody secretion and laser-mediated purification for rapid cloning of highly-secreting producers.
Hanania EG, Fieck A, Stevens J, Bodzin LJ, Palsson BØ, Koller MR. 2005. Automated in situ measurement of cell-specific antibody secretion and laser-mediated purification for rapid cloning of highly-secreting producers.. Biotechnology and bioengineering. 91(7):872-6.
-
Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays.
Herring CD, Raffaelle M, Allen TE, Kanin EI, Landick R, Ansari AZ, Palsson BØ. 2005. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays.. Journal of bacteriology. 187(17):6166-74.
-
The econometrics of evolution.
Fong S, Joyce AR, Palsson BØ. 2005. The econometrics of evolution.. Nature chemical biology. 1(4):191-2.
-
Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states.
Fong S, Joyce AR, Palsson BØ. 2005. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states.. Genome research. 15(10):1365-72.
-
Candidate metabolic network states in human mitochondria. Impact of diabetes, ischemia, and diet.
Thiele I, Price ND, Vo TD, Palsson BØ. 2005. Candidate metabolic network states in human mitochondria. Impact of diabetes, ischemia, and diet.. The Journal of biological chemistry. 280(12):11683-95.
-
k-Cone analysis: determining all candidate values for kinetic parameters on a network scale.
Famili I, Mahadevan R, Palsson BØ. 2005. k-Cone analysis: determining all candidate values for kinetic parameters on a network scale.. Biophysical journal. 88(3):1616-25.
-
Properties of metabolic networks: structure versus function.
Mahadevan R, Palsson BØ. 2005. Properties of metabolic networks: structure versus function.. Biophysical journal. 88(1):L07-9.
-
Reconstruction of cellular signalling networks and analysis of their properties.
Papin JA, Hunter T, Palsson BØ, Subramaniam S. 2005. Reconstruction of cellular signalling networks and analysis of their properties.. Nature reviews. Molecular cell biology. 6(2):99-111.
-
The global transcriptional regulatory network for metabolism in Escherichia coli exhibits few dominant functional states.
Barrett CL, Herring CD, Reed JL, Palsson BØ. 2005. The global transcriptional regulatory network for metabolism in Escherichia coli exhibits few dominant functional states.. Proceedings of the National Academy of Sciences of the United States of America. 102(52):19103-8.
-
Expanded metabolic reconstruction of Helicobacter pylori (iIT341 GSM/GPR): an in silico genome-scale characterization of single- and double-deletion mutants.
Thiele I, Vo TD, Price ND, Palsson BØ. 2005. Expanded metabolic reconstruction of Helicobacter pylori (iIT341 GSM/GPR): an in silico genome-scale characterization of single- and double-deletion mutants.. Journal of bacteriology. 187(16):5818-30.
-
Expa: a program for calculating extreme pathways in biochemical reaction networks.
Bell SL, Palsson BØ. 2005. Expa: a program for calculating extreme pathways in biochemical reaction networks.. Bioinformatics (Oxford, England). 21(8):1739-40.
-
Untangling the web of functional and physical interactions in yeast.
Herrgard MJ, Palsson BØ. 2005. Untangling the web of functional and physical interactions in yeast.. Journal of biology. 4(2):5.
-
Genome-scale reconstruction of the metabolic network in Staphylococcus aureus N315: an initial draft to the two-dimensional annotation.
Becker SA, Palsson BØ. 2005. Genome-scale reconstruction of the metabolic network in Staphylococcus aureus N315: an initial draft to the two-dimensional annotation.. BMC microbiology. 5:8.
-
Integrated analysis of metabolic phenotypes in Saccharomyces cerevisiae.
Duarte NC, Palsson BØ, Fu P. 2004. Integrated analysis of metabolic phenotypes in Saccharomyces cerevisiae.. BMC genomics. 5:63.
-
Reconstruction and functional characterization of the human mitochondrial metabolic network based on proteomic and biochemical data.
Vo TD, Greenberg HJ, Palsson BØ. 2004. Reconstruction and functional characterization of the human mitochondrial metabolic network based on proteomic and biochemical data.. The Journal of biological chemistry. 279(38):39532-40.
-
Genome-scale in silico models of E. coli have multiple equivalent phenotypic states: assessment of correlated reaction subsets that comprise network states.
Reed JL, Palsson BØ. 2004. Genome-scale in silico models of E. coli have multiple equivalent phenotypic states: assessment of correlated reaction subsets that comprise network states.. Genome research. 14(9):1797-805.
-
High-throughput laser-mediated in situ cell purification with high purity and yield.
Koller MR, Hanania EG, Stevens J, Eisfeld TM, Sasaki GC, Fieck A, Palsson BØ. 2004. High-throughput laser-mediated in situ cell purification with high purity and yield.. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 61(2):153-61.
-
Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes.
Fong S, Palsson BØ. 2004. Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes.. Nature genetics. 36(10):1056-8.
-
Uniform sampling of steady-state flux spaces: means to design experiments and to interpret enzymopathies.
Price ND, Schellenberger J, Palsson BØ. 2004. Uniform sampling of steady-state flux spaces: means to design experiments and to interpret enzymopathies.. Biophysical journal. 87(4):2172-86.
-
The evolution of molecular biology into systems biology.
Westerhoff HV, Palsson BØ. 2004. The evolution of molecular biology into systems biology.. Nature biotechnology. 22(10):1249-52.
-
Genome-scale models of microbial cells: evaluating the consequences of constraints.
Price ND, Reed JL, Palsson BØ. 2004. Genome-scale models of microbial cells: evaluating the consequences of constraints.. Nature reviews. Microbiology. 2(11):886-97.
-
Integrating high-throughput and computational data elucidates bacterial networks.
Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BØ. 2004. Integrating high-throughput and computational data elucidates bacterial networks.. Nature. 429(6987):92-6.
-
Using metabolic flux data to further constrain the metabolic solution space and predict internal flux patterns: the Escherichia coli spectrum.
Wiback SJ, Mahadevan R, Palsson BØ. 2004. Using metabolic flux data to further constrain the metabolic solution space and predict internal flux patterns: the Escherichia coli spectrum.. Biotechnology and bioengineering. 86(3):317-31.
-
Topological analysis of mass-balanced signaling networks: a framework to obtain network properties including crosstalk.
Papin JA, Palsson BØ. 2004. Topological analysis of mass-balanced signaling networks: a framework to obtain network properties including crosstalk.. Journal of theoretical biology. 227(2):283-97.PubMed Google Scholar
-
Monte Carlo sampling can be used to determine the size and shape of the steady-state flux space.
Wiback SJ, Famili I, Greenberg HJ, Palsson BØ. 2004. Monte Carlo sampling can be used to determine the size and shape of the steady-state flux space.. Journal of theoretical biology. 228(4):437-47.
-
Flagellar biosynthesis in silico: building quantitative models of regulatory networks.
Herrgard MJ, Palsson BØ. 2004. Flagellar biosynthesis in silico: building quantitative models of regulatory networks.. Cell. 117(6):689-90.
-
Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model.
Duarte NC, Herrgard MJ, Palsson BØ. 2004. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model.. Genome research. 14(7):1298-309.
-
The JAK-STAT signaling network in the human B-cell: an extreme signaling pathway analysis.
Papin JA, Palsson BØ. 2004. The JAK-STAT signaling network in the human B-cell: an extreme signaling pathway analysis.. Biophysical journal. 87(1):37-46.
-
In Silico Metabolic Model and Protein Expression of Haemophilus influenzae Strain Rd KW20 in Rich Medium.
Raghunathan A, Price ND, Galperin MY, Makarova KS, Purvine S, Picone AF, Cherny T, Xie T, Reilly TJ, Munson R et al.. 2004. In Silico Metabolic Model and Protein Expression of Haemophilus influenzae Strain Rd KW20 in Rich Medium.. Omics : a journal of integrative biology. 8(1):25-41.
-
In silico biotechnology. Era of reconstruction and interrogation.
Palsson BØ. 2004. In silico biotechnology. Era of reconstruction and interrogation.. Current opinion in biotechnology. 15(1):50-1.
-
Reconstruction of microbial transcriptional regulatory networks.
Herrgard MJ, Covert MW, Palsson BØ. 2004. Reconstruction of microbial transcriptional regulatory networks.. Current opinion in biotechnology. 15(1):70-7.
-
Hierarchical thinking in network biology: the unbiased modularization of biochemical networks.
Papin JA, Reed JL, Palsson BØ. 2004. Hierarchical thinking in network biology: the unbiased modularization of biochemical networks.. Trends in biochemical sciences. 29(12):641-7.
-
High-throughput mutation detection underlying adaptive evolution of Escherichia coli-K12.
Honisch C, Raghunathan A, Cantor CR, Palsson BØ, van den Boom D. 2004. High-throughput mutation detection underlying adaptive evolution of Escherichia coli-K12.. Genome research. 14(12):2495-502.
-
Comparison of network-based pathway analysis methods.
Papin JA, Stelling J, Price ND, Klamt S, Schuster S, Palsson BØ. 2004. Comparison of network-based pathway analysis methods.. Trends in biotechnology. 22(8):400-5.
-
Systemic metabolic reactions are obtained by singular value decomposition of genome-scale stoichiometric matrices.
Famili I, Palsson BØ. 2003. Systemic metabolic reactions are obtained by singular value decomposition of genome-scale stoichiometric matrices.. Journal of theoretical biology. 224(1):87-96.
-
Reconstructing metabolic flux vectors from extreme pathways: defining the alpha-spectrum.
Wiback SJ, Mahadevan R, Palsson BØ. 2003. Reconstructing metabolic flux vectors from extreme pathways: defining the alpha-spectrum.. Journal of theoretical biology. 224(3):313-24.
-
Network-based analysis of metabolic regulation in the human red blood cell.
Price ND, Reed JL, Papin JA, Wiback SJ, Palsson BØ. 2003. Network-based analysis of metabolic regulation in the human red blood cell.. Journal of theoretical biology. 225(2):185-94.
-
Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network.
Famili I, Forster J, Nielsen J, Palsson BØ. 2003. Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network.. Proceedings of the National Academy of Sciences of the United States of America. 100(23):13134-9.
-
Reconciling gene expression data with known genome-scale regulatory network structures.
Herrgard MJ, Covert MW, Palsson BØ. 2003. Reconciling gene expression data with known genome-scale regulatory network structures.. Genome research. 13(11):2423-34.PubMed Google Scholar
-
Genome-scale analysis of the uses of the Escherichia coli genome: model-driven analysis of heterogeneous data sets.
Allen TE, Herrgard MJ, Liu M, Qiu Y, Glasner JD, Blattner FR, Palsson BØ. 2003. Genome-scale analysis of the uses of the Escherichia coli genome: model-driven analysis of heterogeneous data sets.. Journal of bacteriology. 185(21):6392-9.
-
Description and interpretation of adaptive evolution of Escherichia coli K-12 MG1655 by using a genome-scale in silico metabolic model.
Fong S, Marciniak JY, Palsson BØ. 2003. Description and interpretation of adaptive evolution of Escherichia coli K-12 MG1655 by using a genome-scale in silico metabolic model.. Journal of bacteriology. 185(21):6400-8.
-
Thirteen years of building constraint-based in silico models of Escherichia coli.
Reed JL, Palsson BØ. 2003. Thirteen years of building constraint-based in silico models of Escherichia coli.. Journal of bacteriology. 185(9):2692-9.
-
Development of network-based pathway definitions: the need to analyze real metabolic networks.
Palsson BØ, Price ND, Papin JA. 2003. Development of network-based pathway definitions: the need to analyze real metabolic networks.. Trends in biotechnology. 21(5):195-8.
-
Metabolic pathways in the post-genome era.
Papin JA, Price ND, Wiback SJ, Fell DA, Palsson BØ. 2003. Metabolic pathways in the post-genome era.. Trends in biochemical sciences. 28(5):250-8.
-
Scalable method to determine mutations that occur during adaptive evolution of Escherichia coli.
Raghunathan A, Palsson BØ. 2003. Scalable method to determine mutations that occur during adaptive evolution of Escherichia coli.. Biotechnology letters. 25(5):435-41.PubMed Google Scholar
-
Large-scale evaluation of in silico gene deletions in Saccharomyces cerevisiae.
Förster J, Famili I, Palsson BØ, Nielsen J. 2003. Large-scale evaluation of in silico gene deletions in Saccharomyces cerevisiae.. Omics : a journal of integrative biology. 7(2):193-202.PubMed Google Scholar
-
The convex basis of the left null space of the stoichiometric matrix leads to the definition of metabolically meaningful pools.
Famili I, Palsson BØ. 2003. The convex basis of the left null space of the stoichiometric matrix leads to the definition of metabolically meaningful pools.. Biophysical journal. 85(1):16-26.
-
Sequence-based analysis of metabolic demands for protein synthesis in prokaryotes.
Allen TE, Palsson BØ. 2003. Sequence-based analysis of metabolic demands for protein synthesis in prokaryotes.. Journal of theoretical biology. 220(1):1-18.
-
Analysis of metabolic capabilities using singular value decomposition of extreme pathway matrices.
Price ND, Reed JL, Papin JA, Famili I, Palsson BØ. 2003. Analysis of metabolic capabilities using singular value decomposition of extreme pathway matrices.. Biophysical journal. 84(2 Pt 1):794-804.
-
Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network.
Förster J, Famili I, Fu P, Palsson BØ, Nielsen J. 2003. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network.. Genome research. 13(2):244-53.
-
Identifying constraints that govern cell behavior: a key to converting conceptual to computational models in biology?
Covert MW, Famili I, Palsson BØ. 2003. Identifying constraints that govern cell behavior: a key to converting conceptual to computational models in biology? Biotechnology and bioengineering. 84(7):763-72.PubMed Google Scholar
-
Initial proteome analysis of model microorganism Haemophilus influenzae strain Rd KW20.
Kolker E, Purvine S, Galperin MY, Stolyar S, Goodlett DR, Nesvizhskii AI, Keller A, Xie T, Eng JK, Yi E et al.. 2003. Initial proteome analysis of model microorganism Haemophilus influenzae strain Rd KW20.. Journal of bacteriology. 185(15):4593-602.
-
Constraints-based models: regulation of gene expression reduces the steady-state solution space.
Covert MW, Palsson BØ. 2003. Constraints-based models: regulation of gene expression reduces the steady-state solution space.. Journal of theoretical biology. 221(3):309-25.
-
Genome-scale microbial in silico models: the constraints-based approach.
Price ND, Papin JA, Schilling CH, Palsson BØ. 2003. Genome-scale microbial in silico models: the constraints-based approach.. Trends in biotechnology. 21(4):162-9.
-
Quantitative analysis of Escherichia coli metabolic phenotypes within the context of phenotypic phase planes.
Ibarra RU, Fu P, Palsson BØ, DiTonno JR, Edwards JS. 2003. Quantitative analysis of Escherichia coli metabolic phenotypes within the context of phenotypic phase planes.. Journal of molecular microbiology and biotechnology. 6(2):101-8.PubMed Google Scholar
-
An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR).
Reed JL, Vo TD, Schilling CH, Palsson BØ. 2003. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR).. Genome biology. 4(9):R54.
-
Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth.
Ibarra RU, Edwards JS, Palsson BØ. 2002. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth.. Nature. 420(6912):186-9.
-
Extreme pathways and Kirchhoff's second law.
Price ND, Famili I, Beard DA, Palsson BØ. 2002. Extreme pathways and Kirchhoff's second law.. Biophysical journal. 83(5):2879-82.
-
In silico model-driven assessment of the effects of single nucleotide polymorphisms (SNPs) on human red blood cell metabolism.
Jamshidi N, Wiback SJ, Palsson BØ. 2002. In silico model-driven assessment of the effects of single nucleotide polymorphisms (SNPs) on human red blood cell metabolism.. Genome research. 12(11):1687-92.
-
Determination of redundancy and systems properties of the metabolic network of Helicobacter pylori using genome-scale extreme pathway analysis.
Price ND, Papin JA, Palsson BØ. 2002. Determination of redundancy and systems properties of the metabolic network of Helicobacter pylori using genome-scale extreme pathway analysis.. Genome research. 12(5):760-9.
-
Murine Sca-1(+)/Lin(-) cells and human KG1a cells exhibit multiple pseudopod morphologies during migration.
Francis K, Palsson BØ, Donahue J, Fong S, Carrier E. 2002. Murine Sca-1(+)/Lin(-) cells and human KG1a cells exhibit multiple pseudopod morphologies during migration.. Experimental hematology. 30(5):460-3.
-
The genome-scale metabolic extreme pathway structure in Haemophilus influenzae shows significant network redundancy.
Papin JA, Price ND, Edwards JS, Palsson BØ. 2002. The genome-scale metabolic extreme pathway structure in Haemophilus influenzae shows significant network redundancy.. Journal of theoretical biology. 215(1):67-82.
-
Metabolic modelling of microbes: the flux-balance approach.
Edwards JS, Covert MW, Palsson BØ. 2002. Metabolic modelling of microbes: the flux-balance approach.. Environmental microbiology. 4(3):133-40.
-
Characterizing the metabolic phenotype: a phenotype phase plane analysis.
Edwards JS, Ramakrishna R, Palsson BØ. 2002. Characterizing the metabolic phenotype: a phenotype phase plane analysis.. Biotechnology and bioengineering. 77(1):27-36.
-
Characterization and efficacy of PKH26 as a probe to study the replication history of the human hematopoietic KG1a progenitor cell line.
Lee G M, Fong S, Oh D J, Francis K, Palsson BØ. 2002. Characterization and efficacy of PKH26 as a probe to study the replication history of the human hematopoietic KG1a progenitor cell line.. In vitro cellular & developmental biology. Animal. 38(2):90-6.
-
Extreme pathway lengths and reaction participation in genome-scale metabolic networks.
Papin JA, Price ND, Palsson BØ. 2002. Extreme pathway lengths and reaction participation in genome-scale metabolic networks.. Genome research. 12(12):1889-900.PubMed Google Scholar
-
Transcriptional regulation in constraints-based metabolic models of Escherichia coli.
Covert MW, Palsson BØ. 2002. Transcriptional regulation in constraints-based metabolic models of Escherichia coli.. The Journal of biological chemistry. 277(31):28058-64.
-
Description and analysis of metabolic connectivity and dynamics in the human red blood cell.
Kauffman KJ, Pajerowski J D, Jamshidi N, Palsson BØ, Edwards JS. 2002. Description and analysis of metabolic connectivity and dynamics in the human red blood cell.. Biophysical journal. 83(2):646-62.
-
Extreme pathway analysis of human red blood cell metabolism.
Wiback SJ, Palsson BØ. 2002. Extreme pathway analysis of human red blood cell metabolism.. Biophysical journal. 83(2):808-18.
-
Genome-scale metabolic model of Helicobacter pylori 26695.
Schilling CH, Covert MW, Famili I, Church GM, Edwards JS, Palsson BØ. 2002. Genome-scale metabolic model of Helicobacter pylori 26695.. Journal of bacteriology. 184(16):4582-93.
-
H. influenzae Consortium: integrative study of H. influenzae-human interactions.
Kolker E, Purvine S, Picone A, Cherny T, Akerley BJ, Munson RS, Palsson BØ, Daines DA, Smith AL. 2002. H. influenzae Consortium: integrative study of H. influenzae-human interactions.. Omics : a journal of integrative biology. 6(4):341-8.
-
Regulation of gene expression in flux balance models of metabolism.
Covert MW, Schilling CH, Palsson BØ. 2001. Regulation of gene expression in flux balance models of metabolism.. Journal of theoretical biology. 213(1):73-88.
-
Dynamic simulation of the human red blood cell metabolic network.
Jamshidi N, Edwards JS, Fahland T, Church GM, Palsson BØ. 2001. Dynamic simulation of the human red blood cell metabolic network.. Bioinformatics (Oxford, England). 17(3):286-7.
-
Flux-balance analysis of mitochondrial energy metabolism: consequences of systemic stoichiometric constraints.
Ramakrishna R, Edwards JS, McCulloch A, Palsson BØ. 2001. Flux-balance analysis of mitochondrial energy metabolism: consequences of systemic stoichiometric constraints.. American journal of physiology. Regulatory, integrative and comparative physiology. 280(3):R695-704.
-
Metabolic modeling of microbial strains in silico.
Covert MW, Schilling CH, Famili I, Edwards JS, Goryanin II, Selkov E, Palsson BØ. 2001. Metabolic modeling of microbial strains in silico.. Trends in biochemical sciences. 26(3):179-86.
-
In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data.
Edwards JS, Ibarra RU, Palsson BØ. 2001. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data.. Nature biotechnology. 19(2):125-30.
-
Robustness analysis of the Escherichia coli metabolic network.
Edwards JS, Palsson BØ. 2000. Robustness analysis of the Escherichia coli metabolic network.. Biotechnology progress. 16(6):927-39.
-
Multiple steady states in kinetic models of red cell metabolism.
Edwards JS, Palsson BØ. 2000. Multiple steady states in kinetic models of red cell metabolism.. Journal of theoretical biology. 207(1):125-7.
-
The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities.
Edwards JS, Palsson BØ. 2000. The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities.. Proceedings of the National Academy of Sciences of the United States of America. 97(10):5528-33.
-
Extension of osmolality-induced podia is observed from fluorescently labeled hematopoietic cell lines in hyperosmotic medium.
Oh DJ, Martinez AR, Lee GM, Francis K, Palsson BØ. 2000. Extension of osmolality-induced podia is observed from fluorescently labeled hematopoietic cell lines in hyperosmotic medium.. Cytometry. 40(2):109-18.
-
Intercellular adhesion can be visualized using fluorescently labeled fibrosarcoma HT1080 cells cocultured with hematopoietic cell lines or CD34(+) enriched human mobilized peripheral blood cells.
Oh DJ, Martinez AR, Lee GM, Francis K, Palsson BØ. 2000. Intercellular adhesion can be visualized using fluorescently labeled fibrosarcoma HT1080 cells cocultured with hematopoietic cell lines or CD34(+) enriched human mobilized peripheral blood cells.. Cytometry. 40(2):119-25.
-
In situ labeling of adherent cells with PKH26.
Lee GM, Fong S, Francis K, Oh DJ, Palsson BØ. 2000. In situ labeling of adherent cells with PKH26.. In vitro cellular & developmental biology. Animal. 36(1):4-6.
-
Theory for the systemic definition of metabolic pathways and their use in interpreting metabolic function from a pathway-oriented perspective.
Schilling CH, Letscher D, Palsson BØ. 2000. Theory for the systemic definition of metabolic pathways and their use in interpreting metabolic function from a pathway-oriented perspective.. Journal of theoretical biology. 203(3):229-48.
-
Assessment of the metabolic capabilities of Haemophilus influenzae Rd through a genome-scale pathway analysis.
Schilling CH, Palsson BØ. 2000. Assessment of the metabolic capabilities of Haemophilus influenzae Rd through a genome-scale pathway analysis.. Journal of theoretical biology. 203(3):249-83.
-
Metabolic flux balance analysis and the in silico analysis of Escherichia coli K-12 gene deletions.
Edwards JS, Palsson BØ. 2000. Metabolic flux balance analysis and the in silico analysis of Escherichia coli K-12 gene deletions.. BMC bioinformatics. 1:1.
-
Combining pathway analysis with flux balance analysis for the comprehensive study of metabolic systems.
Schilling CH, Edwards JS, Letscher D, Palsson BØ. 2000. Combining pathway analysis with flux balance analysis for the comprehensive study of metabolic systems.. Biotechnology and bioengineering. 71(4):286-306.
-
Symmetry of initial cell divisions among primitive hematopoietic progenitors is independent of ontogenic age and regulatory molecules.
Huang S, Law P, Francis K, Palsson BØ, Ho AD. 1999. Symmetry of initial cell divisions among primitive hematopoietic progenitors is independent of ontogenic age and regulatory molecules.. Blood. 94(8):2595-604.
-
Key adhesion molecules are present on long podia extended by hematopoietic cells.
Holloway W, Martinez AR, Oh DJ, Francis K, Ramakrishna R, Palsson BØ. 1999. Key adhesion molecules are present on long podia extended by hematopoietic cells.. Cytometry. 37(3):171-7.
-
Toward metabolic phenomics: analysis of genomic data using flux balances.
Schilling CH, Edwards JS, Palsson BØ. 1999. Toward metabolic phenomics: analysis of genomic data using flux balances.. Biotechnology progress. 15(3):288-95.
-
Metabolic pathway analysis: basic concepts and scientific applications in the post-genomic era.
Schilling CH, Schuster S, Palsson BØ, Heinrich R. 1999. Metabolic pathway analysis: basic concepts and scientific applications in the post-genomic era.. Biotechnology progress. 15(3):296-303.
-
Systems properties of the Haemophilus influenzae Rd metabolic genotype.
Edwards JS, Palsson BØ. 1999. Systems properties of the Haemophilus influenzae Rd metabolic genotype.. The Journal of biological chemistry. 274(25):17410-6.
-
Phototoxicity of the fluorescent membrane dyes PKH2 and PKH26 on the human hematopoietic KG1a progenitor cell line.
Oh DJ, Lee GM, Francis K, Palsson BØ. 1999. Phototoxicity of the fluorescent membrane dyes PKH2 and PKH26 on the human hematopoietic KG1a progenitor cell line.. Cytometry. 36(4):312-8.
-
Elemental balancing of biomass and medium composition enhances growth capacity in high-density Chlorella vulgaris cultures.
Mandalam RK, Palsson BØ. 1998. Elemental balancing of biomass and medium composition enhances growth capacity in high-density Chlorella vulgaris cultures.. Biotechnology and bioengineering. 59(5):605-11.
-
Two new pseudopod morphologies displayed by the human hematopoietic KG1a progenitor cell line and by primary human CD34(+) cells.
Francis K, Ramakrishna R, Holloway W, Palsson BØ. 1998. Two new pseudopod morphologies displayed by the human hematopoietic KG1a progenitor cell line and by primary human CD34(+) cells.. Blood. 92(10):3616-23.
-
Tissue culture surface characteristics influence the expansion of human bone marrow cells.
Koller MR, Palsson MA, Manchel I, Maher RJ, Palsson BØ. 1998. Tissue culture surface characteristics influence the expansion of human bone marrow cells.. Biomaterials. 19(21):1963-72.
-
How will bioinformatics influence metabolic engineering?
Edwards JS, Palsson BØ. 1998. How will bioinformatics influence metabolic engineering? Biotechnology and bioengineering. 58(2-3):162-9.
-
Cell cycle dependence of retroviral transduction: An issue of overlapping time scales.
Andreadis ST, Fuller AO, Palsson BØ. 1998. Cell cycle dependence of retroviral transduction: An issue of overlapping time scales.. Biotechnology and bioengineering. 58(2-3):272-81.
-
The underlying pathway structure of biochemical reaction networks.
Schilling CH, Palsson BØ. 1998. The underlying pathway structure of biochemical reaction networks.. Proceedings of the National Academy of Sciences of the United States of America. 95(8):4193-8.
-
Moloney murine leukemia virus-derived retroviral vectors decay intracellularly with a half-life in the range of 5.5 to 7.5 hours.
Andreadis ST, Brott DA, Fuller AO, Palsson BØ. 1997. Moloney murine leukemia virus-derived retroviral vectors decay intracellularly with a half-life in the range of 5.5 to 7.5 hours.. Journal of virology. 71(10):7541-8.
-
Effective intercellular communication distances are determined by the relative time constants for cyto/chemokine secretion and diffusion.
Francis K, Palsson BØ. 1997. Effective intercellular communication distances are determined by the relative time constants for cyto/chemokine secretion and diffusion.. Proceedings of the National Academy of Sciences of the United States of America. 94(23):12258-62.
-
What lies beyond bioinformatics?
Palsson BØ. 1997. What lies beyond bioinformatics? Nature biotechnology. 15(1):3-4.
-
Coupled effects of polybrene and calf serum on the efficiency of retroviral transduction and the stability of retroviral vectors.
Andreadis ST, Palsson BØ. 1997. Coupled effects of polybrene and calf serum on the efficiency of retroviral transduction and the stability of retroviral vectors.. Human gene therapy. 8(3):285-91.
-
Importance of parenchymal:stromal cell ratio for the ex vivo reconstitution of human hematopoiesis.
Koller MR, Manchel I, Palsson BØ. 1997. Importance of parenchymal:stromal cell ratio for the ex vivo reconstitution of human hematopoiesis.. Stem cells (Dayton, Ohio). 15(4):305-13.
-
Kinetics of retrovirus mediated gene transfer: the importance of intracellular half-life of retroviruses.
Andreadis ST, Palsson BØ. 1996. Kinetics of retrovirus mediated gene transfer: the importance of intracellular half-life of retroviruses.. Journal of theoretical biology. 182(1):1-20.
-
Unilineage model of hematopoiesis predicts self-renewal of stem and progenitor cells based on ex vivo growth data.
Peng CA, Koller MR, Palsson BØ. 1996. Unilineage model of hematopoiesis predicts self-renewal of stem and progenitor cells based on ex vivo growth data.. Biotechnology and bioengineering. 52(1):24-33.
-
flt-3 ligand is more potent than c-kit ligand for the synergistic stimulation of ex vivo hematopoietic cell expansion.
Koller MR, Oxender M, Brott DA, Palsson BØ. 1996. flt-3 ligand is more potent than c-kit ligand for the synergistic stimulation of ex vivo hematopoietic cell expansion.. Journal of hematotherapy. 5(5):449-59.
-
Donor-to-donor variability in the expansion potential of human bone marrow cells is reduced by accessory cells but not by soluble growth factors.
Koller MR, Manchel I, Brott DA, Brott DA. 1996. Donor-to-donor variability in the expansion potential of human bone marrow cells is reduced by accessory cells but not by soluble growth factors.. Experimental hematology. 24(13):1484-93.
-
Determination of specific oxygen uptake rates in human hematopoietic cultures and implications for bioreactor design.
Peng CA, Palsson BØ. 1996. Determination of specific oxygen uptake rates in human hematopoietic cultures and implications for bioreactor design.. Annals of biomedical engineering. 24(3):373-81.
-
Cell growth and differentiation on feeder layers is predicted to be influenced by bioreactor geometry.
Peng CA, Palsson BØ. 1996. Cell growth and differentiation on feeder layers is predicted to be influenced by bioreactor geometry.. Biotechnology and bioengineering. 50(5):479-92.
-
Different measures of human hematopoietic cell culture performance are optimized under vastly different conditions.
Koller MR, Manchel I, Palsson MA, Maher RJ, Palsson BØ. 1996. Different measures of human hematopoietic cell culture performance are optimized under vastly different conditions.. Biotechnology and bioengineering. 50(5):505-13.
-
Membrane adsorption characteristics determine the kinetics of flow-through transductions.
Chuck AS, Palsson BØ. 1996. Membrane adsorption characteristics determine the kinetics of flow-through transductions.. Biotechnology and bioengineering. 51(3):260-70.
-
Retroviral infection is limited by Brownian motion.
Chuck AS, Clarke MF, Palsson BØ. 1996. Retroviral infection is limited by Brownian motion.. Human gene therapy. 7(13):1527-34.
-
Consistent and high rates of gene transfer can be obtained using flow-through transduction over a wide range of retroviral titers.
Chuck AS, Palsson BØ. 1996. Consistent and high rates of gene transfer can be obtained using flow-through transduction over a wide range of retroviral titers.. Human gene therapy. 7(6):743-50.
-
Flow cytometric analysis of human bone marrow perfusion cultures: erythroid development and relationship with burst-forming units-erythroid.
Rogers CE, Bradley MS, Palsson BØ, Koller MR. 1996. Flow cytometric analysis of human bone marrow perfusion cultures: erythroid development and relationship with burst-forming units-erythroid.. Experimental hematology. 24(5):597-604.
-
Long-term culture-initiating cell expansion is dependent on frequent medium exchange combined with stromal and other accessory cell effects.
Koller MR, Palsson MA, Manchel I, Palsson BØ. 1995. Long-term culture-initiating cell expansion is dependent on frequent medium exchange combined with stromal and other accessory cell effects.. Blood. 86(5):1784-93.
-
Retroviral-mediated gene transfer in human bone marrow cells growth in continuous perfusion culture vessels.
Eipers PG, Krauss JC, Palsson BØ, Emerson SG, Todd RF, Clarke MF. 1995. Retroviral-mediated gene transfer in human bone marrow cells growth in continuous perfusion culture vessels.. Blood. 86(10):3754-62.
-
Growth factor consumption and production in perfusion cultures of human bone marrow correlate with specific cell production.
Koller MR, Bradley MS, Palsson BØ. 1995. Growth factor consumption and production in perfusion cultures of human bone marrow correlate with specific cell production.. Experimental hematology. 23(12):1275-83.
-
Bioreactor expansion of human bone marrow: comparison of unprocessed, density-separated, and CD34-enriched cells.
Koller MR, Manchel I, Newsom BS, Palsson MA, Palsson BØ. 1995. Bioreactor expansion of human bone marrow: comparison of unprocessed, density-separated, and CD34-enriched cells.. Journal of hematotherapy. 4(3):159-69.
-
Parametric sensitivity of stoichiometric flux balance models applied to wild-type Escherichia coli metabolism.
Varma A, Palsson BØ. 1995. Parametric sensitivity of stoichiometric flux balance models applied to wild-type Escherichia coli metabolism.. Biotechnology and bioengineering. 45(1):69-79.
-
In vitro expansion of hematopoietic cells for clinical application.
Emerson SG, Palsson BØ, Clarke MF, Silver SM, Adams PT, Koller MR, Van Zant G, Rummel S, Armstrong RD, Maluta J. 1995. In vitro expansion of hematopoietic cells for clinical application.. Cancer treatment and research. 76:215-23.
-
Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110.
Varma A, Palsson BØ. 1994. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110.. Applied and environmental microbiology. 60(10):3724-31.
-
High-density algal photobioreactors using light-emitting diodes.
Lee CG, Palsson BØ. 1994. High-density algal photobioreactors using light-emitting diodes.. Biotechnology and bioengineering. 44(10):1161-7.
-
Preface. Tissue engineering and cell therapies.
Hubbell JA, Palsson BØ, Papoutsakis ET. 1994. Preface. Tissue engineering and cell therapies.. Biotechnology and bioengineering. 43(7):541.
-
Predictions for oxygen supply control to enhance population stability of engineered production strains.
Varma A, Palsson BØ. 1994. Predictions for oxygen supply control to enhance population stability of engineered production strains.. Biotechnology and bioengineering. 43(4):275-85.
-
Frequent harvesting from perfused bone marrow cultures results in increased overall cell and progenitor expansion.
Oh DJ, Koller MR, Palsson BØ. 1994. Frequent harvesting from perfused bone marrow cultures results in increased overall cell and progenitor expansion.. Biotechnology and bioengineering. 44(5):609-16.
-
Preface: Tissue engineering and cell therapies: II.
Hubbell JA, Palsson BØ, Papoutsakis ET. 1994. Preface: Tissue engineering and cell therapies: II.. Biotechnology and bioengineering. 43(8):683.
-
Microencapsulated human bone marrow cultures: a potential culture system for the clonal outgrowth of hematopoietic progenitor cells.
Levee MG, Lee GM, Paek SH, Palsson BØ. 1994. Microencapsulated human bone marrow cultures: a potential culture system for the clonal outgrowth of hematopoietic progenitor cells.. Biotechnology and bioengineering. 43(8):734-9.
-
Monoclonal antibody production using free-suspended and entrapped hybridoma cells.
Lee GM, Palsson BØ. 1994. Monoclonal antibody production using free-suspended and entrapped hybridoma cells.. Biotechnology & genetic engineering reviews. 12:509-33.
-
Enhanced specific antibody productivity of calcium alginate-entrapped hybridoma is cell line-specific.
Lee GM, Kim SJ, Palsson BØ. 1994. Enhanced specific antibody productivity of calcium alginate-entrapped hybridoma is cell line-specific.. Cytotechnology. 16(1):1-15.
-
Review: tissue engineering: reconstitution of human hematopoiesis ex vivo.
Koller MR, Palsson BØ. 1993. Review: tissue engineering: reconstitution of human hematopoiesis ex vivo.. Biotechnology and bioengineering. 42(8):909-30.PubMed Google Scholar
-
Stability of antibody productivity is improved when hybridoma cells are entrapped in calcium alginate beads.
Lee GM, Palsson BØ. 1993. Stability of antibody productivity is improved when hybridoma cells are entrapped in calcium alginate beads.. Biotechnology and bioengineering. 42(9):1131-5.
-
Simultaneous determination of ammonia nitrogen and L-glutamine in bioreactor media using flow injection.
Palsson BØ, Shen BQ, Meyerhoff ME, Trojanowicz M. 1993. Simultaneous determination of ammonia nitrogen and L-glutamine in bioreactor media using flow injection.. The Analyst. 118(11):1361-5.
-
Expansion of human bone marrow progenitor cells in a high cell density continuous perfusion system.
Palsson BØ, Paek SH, Schwartz RM, Palsson MA, Lee GM, Silver S, Emerson SG. 1993. Expansion of human bone marrow progenitor cells in a high cell density continuous perfusion system.. Bio/technology (Nature Publishing Company). 11(3):368-72.
-
Biochemical production capabilities of Escherichia coli.
Varma A, Boesch BW, Palsson BØ. 1993. Biochemical production capabilities of Escherichia coli.. Biotechnology and bioengineering. 42(1):59-73.
-
Loss of antibody productivity is highly reproducible in multiple hybridoma subclones.
Merritt SE, Palsson BØ. 1993. Loss of antibody productivity is highly reproducible in multiple hybridoma subclones.. Biotechnology and bioengineering. 42(2):247-50.
-
Large-scale expansion of human stem and progenitor cells from bone marrow mononuclear cells in continuous perfusion cultures.
Koller MR, Emerson SG, Palsson BØ. 1993. Large-scale expansion of human stem and progenitor cells from bone marrow mononuclear cells in continuous perfusion cultures.. Blood. 82(2):378-84.
-
Cell Culture conditions determine the enhancement of specific monoclonal antibody productivity of calcium alginate-entrapped S3H5/gamma2bA2 hybridoma cells.
Lee GM, Chuck AS, Palsson BØ. 1993. Cell Culture conditions determine the enhancement of specific monoclonal antibody productivity of calcium alginate-entrapped S3H5/gamma2bA2 hybridoma cells.. Biotechnology and bioengineering. 41(3):330-40.
-
Metabolic capabilities of Escherichia coli: I. synthesis of biosynthetic precursors and cofactors.
Varma A, Palsson BØ. 1993. Metabolic capabilities of Escherichia coli: I. synthesis of biosynthetic precursors and cofactors.. Journal of theoretical biology. 165(4):477-502.
-
Stoichiometric interpretation of Escherichia coli glucose catabolism under various oxygenation rates.
Varma A, Boesch BW, Palsson BØ. 1993. Stoichiometric interpretation of Escherichia coli glucose catabolism under various oxygenation rates.. Applied and environmental microbiology. 59(8):2465-73.
-
Model complexity has a significant effect on the numerical value and interpretation of metabolic sensitivity coefficients.
Palsson BØ, Lee ID. 1993. Model complexity has a significant effect on the numerical value and interpretation of metabolic sensitivity coefficients.. Journal of theoretical biology. 161(3):299-315.
-
Simultaneous enzymatic/electrochemical determination of glucose and L-glutamine in hybridoma media by flow-injection analysis.
Meyerhoff ME, Trojanowicz M, Palsson BØ. 1993. Simultaneous enzymatic/electrochemical determination of glucose and L-glutamine in hybridoma media by flow-injection analysis.. Biotechnology and bioengineering. 41(10):964-9.
-
Continuous photoautotrophic cultures of the eukaryotic alga Chlorella vulgaris can exhibit stable oscillatory dynamics.
Javanmardian M, Palsson BØ. 1992. Continuous photoautotrophic cultures of the eukaryotic alga Chlorella vulgaris can exhibit stable oscillatory dynamics.. Biotechnology and bioengineering. 39(5):487-97.
-
Optimal selection of metabolic fluxes for in vivo measurement. I. Development of mathematical methods.
Savinell JM, Palsson BØ. 1992. Optimal selection of metabolic fluxes for in vivo measurement. I. Development of mathematical methods.. Journal of theoretical biology. 155(2):201-14.
-
Optimal selection of metabolic fluxes for in vivo measurement. II. Application to Escherichia coli and hybridoma cell metabolism.
Savinell JM, Palsson BØ. 1992. Optimal selection of metabolic fluxes for in vivo measurement. II. Application to Escherichia coli and hybridoma cell metabolism.. Journal of theoretical biology. 155(2):215-42.
-
Replication of mini-F plasmids during the bacterial division cycle.
Keasling JD, Palsson BØ, Cooper S. 1992. Replication of mini-F plasmids during the bacterial division cycle.. Research in microbiology. 143(6):541-8.
-
Replication of prophage P1 is cell-cycle specific.
Keasling JD, Palsson BØ, Cooper S. 1992. Replication of prophage P1 is cell-cycle specific.. Journal of bacteriology. 174(13):4457-62.
-
A Macintosh software package for simulation of human red blood cell metabolism.
Lee ID, Palsson BØ. 1992. A Macintosh software package for simulation of human red blood cell metabolism.. Computer methods and programs in biomedicine. 38(4):195-226.
