As systems biologists, we seek to understand dynamic cellular responses at the ‘system’, or genome-scale level. We also want to take advantage of cheap sequencing costs and large amounts of publicly available data. To this end, a team in our Genome Analytics subgroup developed iModulon analysis.
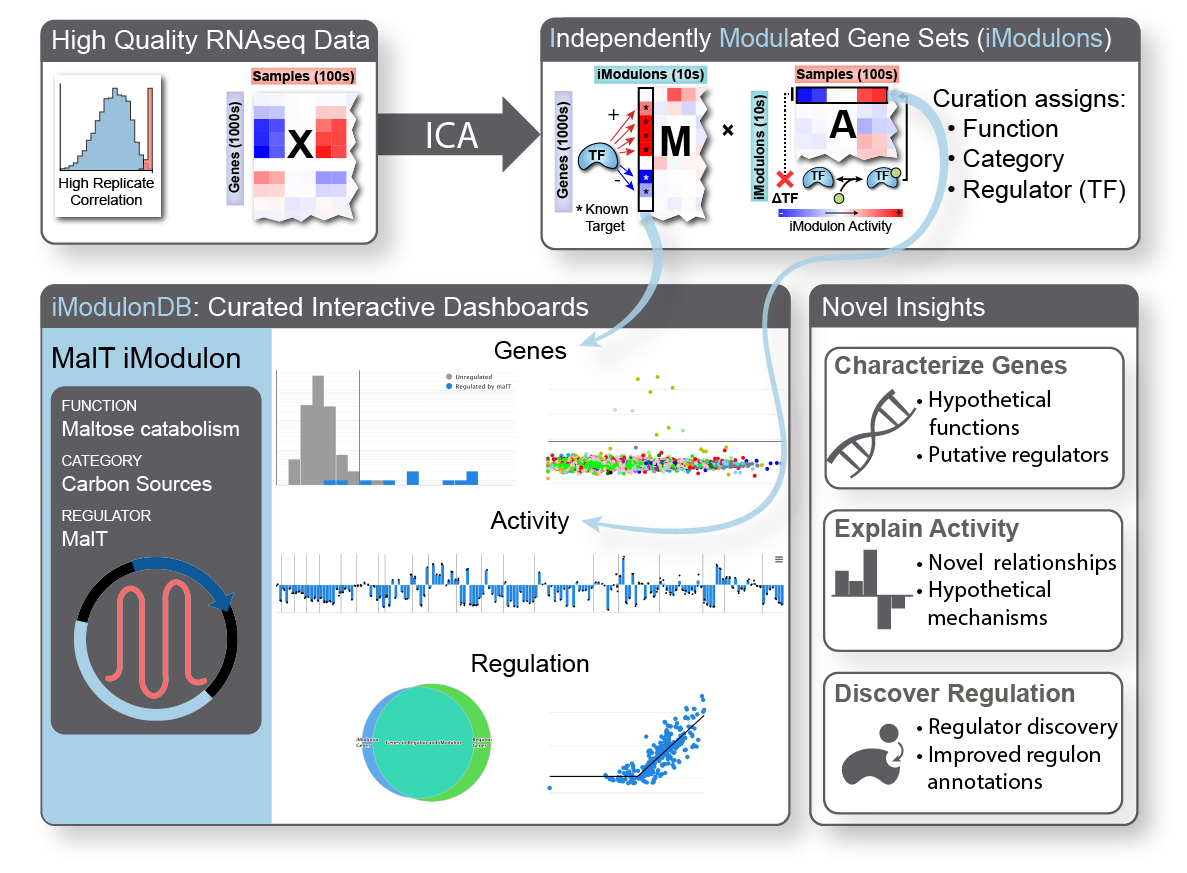
iModulon analysis is the use of a machine learning method called independent component analysis to compute and characterize the transcriptional regulatory network of an organism. We begin with a large compendium of gene expression data, which typically comes from RNA sequencing of cells under a wide range of conditions. We generated a very large E. coli dataset for this purpose, and have also worked with several other organisms. In addition to the in-house data, we download, process, and quality control all available data for organisms of interest from online databases. Once we have a good dataset, we run it through our quantitative pipeline, which computes robust independent components we call iModulons.
iModulons, or independently modulated gene sets, consist of two things: their gene members and activity levels. Each one represents an independent signal, meaning that the set of gene members were co-expressed throughout the dataset. The activity level tells us how active that set of genes is in each sample. Typically, biologists seeking to understand co-regulated gene sets will build ‘regulons’ from the bottom-up using a variety of experiments; instead, we generate analogous structures from the top-down using data alone. Also, typical approaches for comparing conditions in transcriptomic data involve parsing through hundreds or thousands of differentially expressed genes – our method allows us to look at only dozens of differential iModulon activities, finally making it tractable to understand nearly all of the gene expression reallocation that cells perform.
iModulons usually represent the effects of transcription factors and other regulatory elements, allowing us to directly study the transcriptional regulatory network from a new perspective. They can highlight important uncharacterized genes for further study or be used to discover transcription factors. They could contain biosynthetic gene clusters or describe the strongest effects of master regulators. They can be used to break down complex processes like sporulation and competency. They provide new insights into antibiotic resistance mechanisms and the regulation of virulence in pathogens. They also capture effects of small changes to the genome, such as those which result from laboratory evolution. We believe that they will become an increasingly powerful tool in the 2020’s as they continue to be applied to more data.
Current efforts and opportunities in the SBRG around iModulons include:
- Applying them to learn about new organisms
- Studying how they evolve by comparing across related or distant species
- Using them in conjunction with other technologies such as proteomics and metabolomics to gain multi-omic understanding, and
- Using them as modular building blocks to engineer cellular factories
- Continuously developing our online database
To learn more about iModulons, browse all published and curated data, and find links to all relevant publications, see iModulonDB.org.
iModulon efforts are part of the Genome Analytics program at the SBRG, which is led and managed by Kevin Rychel. Kevin is a senior PhD student in the Bioengineering department at UCSD. The Genome Analytics group seeks to develop databases like iModulonDB as well as scientific use cases and workflows which advance basic science, promote collaboration, and support genome design efforts.
Further Reading
- All relevant publications listed in iModulonDB about page here
- Sastry, A.V., Gao, Y., Szubin, R. et al. The Escherichia coli transcriptome mostly consists of independently regulated modules. Nat Commun 10, 5536 (2019).
- Rychel, K., Sastry, A.V., Palsson, B.O. Machine learning uncovers independently regulated modules in the Bacillus subtilis transcriptome. Nat Commun 11, 6338 (2020).
- Poudel, S., Tsunemoto, H., Seif, Y. et al. Revealing 29 sets of independently modulated genes in Staphylococcus aureus, their regulators and role in key physiological responses. PNAS 117 (29) 17228-17239 (2020).
- Rychel, K., Decker, K., Sastry, A.V. et al. iModulonDB: a knowledgebase of microbial transcriptional regulation derived from machine learning. Nucleic Acids Research 49, D112 (2021).