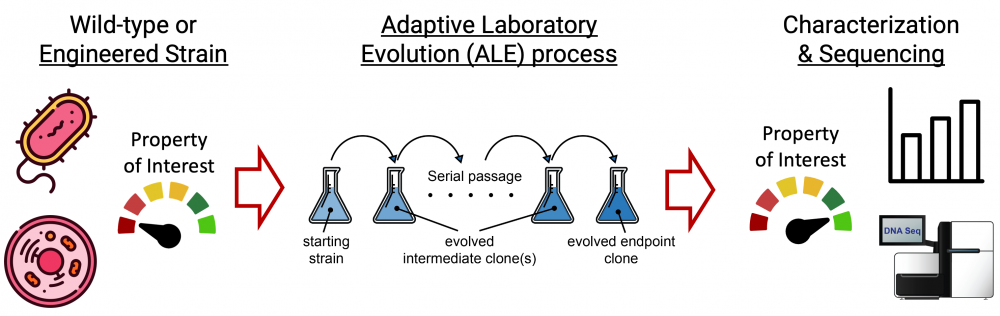
ALE harnesses biology to self-optimize by natural (or induced) mutations through selection. We leverage this process to obtain beneficial strains and mutations.
The Adaptive Laboratory Evolution (ALE) Program is an integral part of the SBRG and harnesses the power of biology to self-optimize under selective pressures and applies this concept to obtain strains with useful phenotypes. Such phenotypes are then linked to genotype and used for discovery, as well as industrial and biomedical applications [1].
With the rise of whole genome sequencing, automation, and bioinformatics, we have built a powerful comprehensive platform to go from a selectable feature or concept to a fully characterized strain with desirable features and a defined and interpreted genotype [2] [3]. We achieve results via high-throughput experimentation and detailed experimental design to drive projects to successful outcomes [4]. We have applied the automated ALE approach to over 25 different bacterial and fungal species and have a dynamic setup to meet the needs of a given application. Our lab also goes beyond the results from a given project by aggregating and organizing mutational data into a quarriable database of causal mutations from laboratory evolution experiments [5]. This approach enables synthetic biology and de novo design of desirable phenotypes as well as providing context for new projects and mutations [6].
The major features of our platform include:
- Robotics and Automation
- Hardware with multiple iterations to enable robustness and flexibility
- Process Control Software for tight experimental control
- Suites of Modules for different Section Pressures
- Algorithms to optimize start up and termination criteria
- True high-throughput evolutionary experimentation with large population sizes and individually-addressable experiments
- Bioinformatics
- Mutation database for rapid dissemination and interpretation, ALEdb
- Cloud-based mutation calling pipeline with QC
- Genome-scale models to predict and interpret evolution outcomes
- Experience and Knowhow
- Several successful use cases and applications
- Standard Operating Procedures (SOPs) followed for all actions
- Strain phenotyping and omics data generation
- Leadership of over 70 peer-reviewed publications
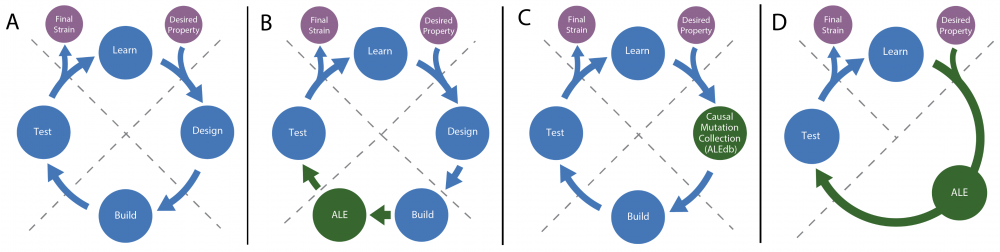
ALE for use in the Design, Build, Test, Learn cycle. A) The typical Design, Build, Test, Learn cycle used in metabolic engineering to generate a strain with a desired property. B) Augmentation of the cycle where ALE is included in the Build step to rescue a strain that has decreased fitness due to a perturbation, or to optimize a strain after removal or addition of genetic content. C) Augmentation of the cycle where a collection of mutations (e.g., ALEdb) associated to a particular phenotype is leveraged for the Design step D) Augmentation of the cycle where ALE can be used to completely replace the Design and Build steps and a desirable strain is acquired directly from ALE when a phenotype can be tied to selection without engineering.
The ALE program in the SBRG is led and managed by Dr. Adam Feist who is an independent PI and Research Scientist in the Bioengineering Department at UCSD. Dr. Feist and SBRG PI Professor Palsson have a nearly 20-year relationship of working both collaboratively and independently on various funded projects to achieve results and educate trainees.
Citations/Further Reading
- Sandberg TE, Salazar MJ, Weng LL, Palsson BO, Feist AM. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab Eng. 2019.
- Lim HG, Eng T, Banerjee D, Alarcon G, Lau AK, Park M-R, et al. Generation of Pseudomonas putida KT2440 Strains with Efficient Utilization of Xylose and Galactose via Adaptive Laboratory Evolution. ACS Sustainable Chem Eng. 2021;9: 11512–11523.
- Anand A, Patel A, Chen K, Olson CA, Phaneuf PV, Lamoureux C, et al. Laboratory evolution of synthetic electron transport system variants reveals a larger metabolic respiratory system and its plasticity. Nat Commun. 2022;13: 3682.
- LaCroix RA, Palsson BO, Feist AM. A Model for Designing Adaptive Laboratory Evolution Experiments. Appl Environ Microbiol. 2017;83.
- Phaneuf PV, Gosting D, Palsson BO, Feist AM. ALEdb 1.0: a database of mutations from adaptive laboratory evolution experimentation. Nucleic Acids Res. 2018.
- Phaneuf PV, Zielinski DC, Yurkovich JT, Johnsen J, Szubin R, Yang L, et al. Escherichia coli Data-Driven Strain Design Using Aggregated Adaptive Laboratory Evolution Mutational Data. ACS Synth Biol. 2021.